When the CRISPR-cas9 system was first discovered it promised to make genome editing easier and cheaper than ever before. And this is partly true: simple knock-outs modifications can be achieved much more easily using CRISPR-cas9 and it is more user-friendly than the genome editing technologies previously available.
However, complicated edits — such as multiplex modifications and large knock-ins — are more difficult. For challenging CRISPR projects, typical efficiency is 0.04% meaning less than one percent of cells are successfully edited. Some cell types are also difficult to edit using CRISPR — such as induced pluripotent stem cells (iPSCs) and primary cells. Because these experiments have such a poor success rate it is harder to identify successfully modified clones.
Researchers at GenAhead Bio have created a more sensitive CRISPR screening using digital droplet PCR (ddPCR) in order to improve the success rate of challenging genome editing projects. In this article, Dr Tsukasa Sugo explains how ddPCR can be used to achieve complicated genome edits at a 97% success rate.
How does digital PCR work?
The key difference between conventional PCR and ddPCR is the number of reactions that take place. During conventional PCR, the sample DNA is amplified in a single reaction. In ddPCR, the sample DNA sample is partitioned into thousands of droplets, so thousands of PCR reactions occur simultaneously. By increasing the number of individual reactions, screening sensitivity is increased and it is possible to detect very low copy numbers of cells.
What are single base-pair changes?
Single base-pair changes, or polymorphisms (SNPs), are tiny mutations that affect one base pair of the DNA. Over 60% of inherited diseases are caused by SNPs; they also contribute to individual sensitivity to medicines. Because these mutations are so small (affecting single base pairs) it is difficult to induce them using conventional genome editing techniques, and with CRISPR knock-in success rate is below 0.05%.
What are induced pluripotent stem cells?
iPSCs are a type of stem cell created from differentiated adult cells. Like their embryonic counterparts, they can differentiate into any cell type, however, they are not created from fetal tissue. There are many benefits to using iPSCs in medicine and research, such as the ability to create different cell types with the same genetic background. iPSCs are particularly advantageous in drug development, especially for diseases where primary tissue is hard to obtain, such as neurodegenerative conditions.
Improving knock-in efficacy with droplet digital PCR
In a 2014 study, Miyaoka et al introduced 30 different SNPs in iPSCs1, achieving a knock-in efficacy of 0.04%. As conventional PCR is not sensitive enough to detect a 0.04% knock-in rate, more advanced screening methods were needed for clonal selection. Following analysis with ddPCR, researchers counted the number of positive wells containing their desired mutation, calculating how many cells in the original mixture had been successfully edited using CRISPR. By using ddPCR, just 11 clones were required for detection.
Without ddPCR, Miyaoka et al would have had to seed 3,000 cells per well during the first round of genome editing, then expanded these cells to achieve a knock-in percentage of 0.73% — repeating this process again would achieve a knock-in percentage of 6.8%. By using ddPCR the process was much less laborious and streamlined.
Using CRISPR-SNIPER to improve genome editing efficiency
The sensitivity of ddPCR can be improved even further with SNIPER analysis: a multi-colored ddPCR screen developed by researchers at GenAhead Bio. SNIPER stands for "specification of newly integrated positions and exclusion of random integration", and is useful for detecting more challenging CRISPR edits such as bi-allelic knock-ins, multiplex knock-outs, and SNPs. By using this multi-colored screen and optimizing the culture conditions, the likely success of a genome editing experiment can be determined in just two weeks.
Example: Creating an Alzheimer's disease model using CRISPR-SNIPER
In this case study, we'll give you an example of how to prepare an Alzheimer's disease (AD) model using CRISPR-SNIPER. One of the hallmarks of AD is a high Aβ42/Aβ40 ratio caused by mutations in the presenilin (PSN) gene, such as proline-117-leucine (P117L). We wanted to create cells containing this mutation using CRISPR SNIPER.
Finding 1: Genome editing efficiency was largely dependent on insert or DNA type
Initially, we had some success with 300 nanograms of an ssDNA insert. Increasing the amount of DNA insert doubled the knock-in efficacy, with optimal efficacy achieved using a long 2kbp insert (figure 1).
| A | 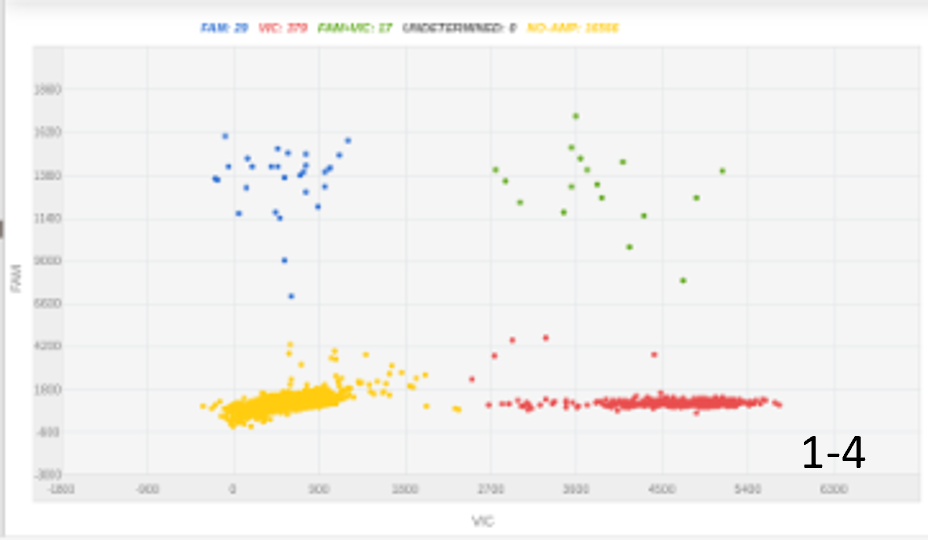 |
B | 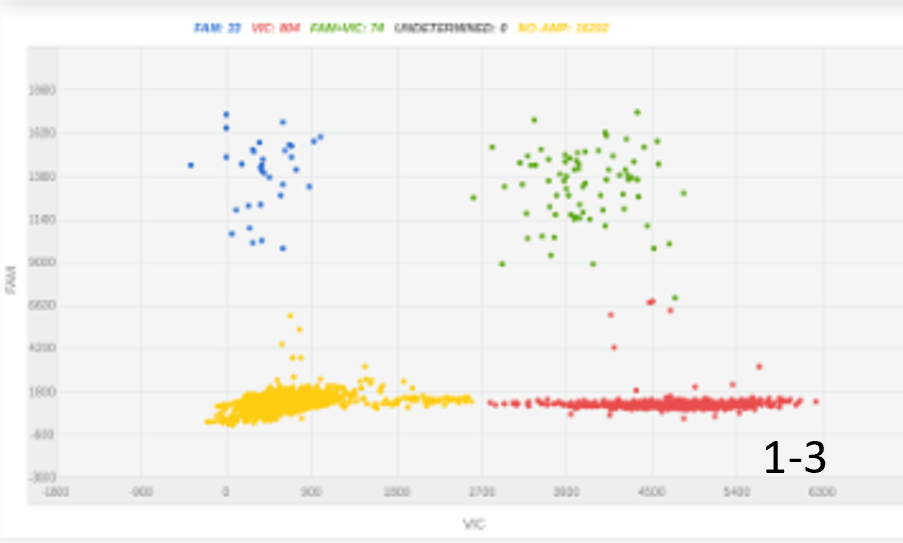 |
C | 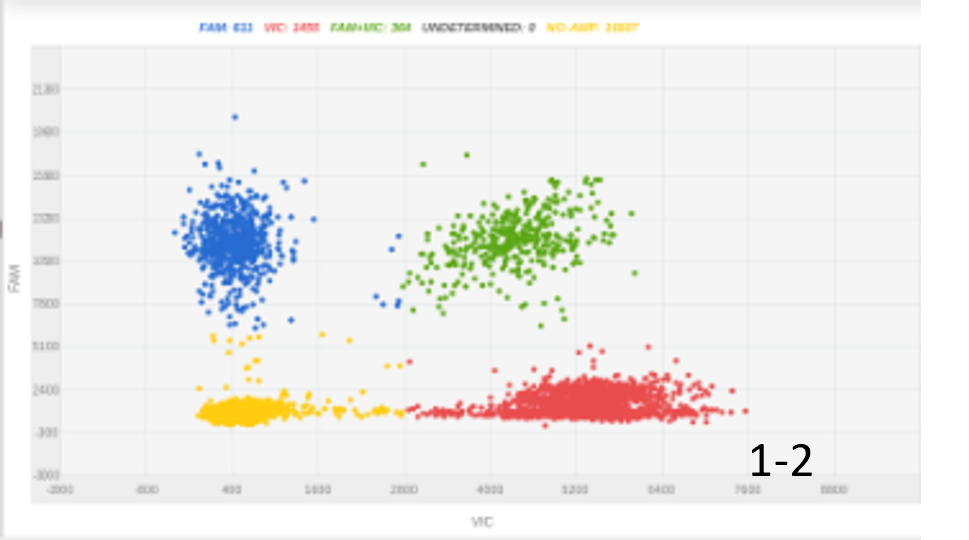 |
Figure 1: The amount of DNA insert and its length were key for optimizing knock-in percentage. A, 100 nucleotides ssODN, 300ng, 4% KI rate; B, 100 nucleotides ssODN, 1000ng, 8% KI rate; C, 2000 nucleotides, 450 ng, 20% KI rate.
Finding 2: Successful identification of inserted molecules and pick-up
Under the optimized genome editing conditions, we then successfully isolated the hetero-mutant and homo-mutant cells. The mutated iPSCs were then differentiated into neurons; results in figure 2 show a nice correlation between genotype and clinical pathology (Aβ42/Aβ40 ratio).
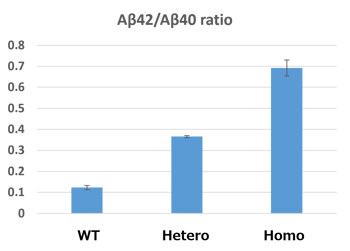
Figure 2: The ratio of Aβ42/Aβ40 in hetero- and homo-mutant neurons differentiated from iPSCs.
Example: Using GFP to track cardiac differentiation
Fluorescent tags, such as green fluorescence protein (GFP), are commonly used to indicate the expression of a specific protein. In iPSC research, fluorescent tagging is useful for optimizing differentiation protocols, screening differentiating compounds with a compound library2, and detecting differentiated cells in organoids. Figure 3 shows a knock-in experiment where GFP was fused to differentiation marker H using CRISPR-SNIPER at a very high knock-in rate (34%).
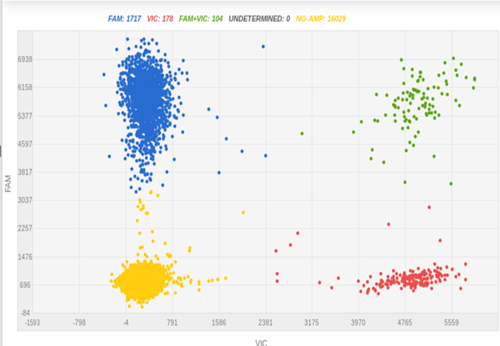
Figure 3: Before iPSC pick-up, we checked the GFP knock-in efficiency in bulk culture using SNIPER, which was 34%.
Example: Transfecting primary cells
While most knock-out edits are easily achieved using CRISPR some cell types are challenging to transfect. Figure 4 shows a knockout experiment where CRISPR-SNIPER was used to achieve a CD4+ specific gRNA knockout from primary T cells. Following knockout, CD4+ expression was measured by flow cytometry, estimating a very high knockout efficacy of 95%.
A
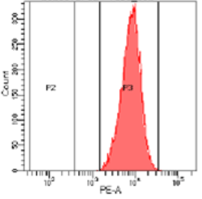
B
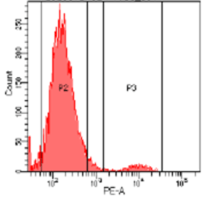
Figure 4: Flow cytometry results. T cells population containing CD4+ marker before (A, 100%) and after (B, 5%) knockout using CRISPR-SNIPER.
CRISPR-SNIPER Genome editing services
CRISPR-SNIPER is available as a genome editing service from GenAhead Bio. The service is divided into two phases:
- Design and feasibility (Basic Package)
genome editing tools are prepared, cutting efficiency of the gRNA optimized, and transfection is performed under six conditions. - Scheduled cloning and QC (Full Package)
If you are satisfied with the knock-in percentage achieved in the design and feasibility phase, we can proceed to QC and expansion of your desired mutant.
If you are interested in discussing a CRISPR-SNIPER study, please make an inquiry through our contact page or visit the CRISPR-SNIPER section of our website to learn more.
Further reading
- Miyaoka et al. Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nature Methods 11:3 pp291-293 (2014)
- Hartogh et al. Concise Review: Fluorescent Reporters in Human Pluripotent Stem Cells: Contributions to Cardiac Differentiation and Their Applications in Cardiac Disease and Toxicity. Stem Cells 34:1 pp13-26 (2016)
Note: The modified cells are developed, manufactured or supplied by GenAhead Bio Inc. under license from ERS Genomics Limited.



.jpg?width=1000&height=562&name=Untitled%20design%20(10).jpg)



