In recent times, interest in the therapeutic use of cells derived from induced pluripotent stem cells (iPSCs) has grown exponentially. These iPSC-derived therapies are currently being developed for the treatment of numerous diseases, including amyotrophic lateral sclerosis (ALS) and corneal epithelial stem cell deficiency.
However, the risk of immunological mismatch between the therapeutic cells and their recipient may limit the use of iPSC therapies clinically. Like transplant rejections, iPSC therapies could face similar issues due to donor-to-recipient mismatches, stimulating an immunogenic response. This response reduces the in vivo survival of the transplanted cells and their efficacy and is mediated at a molecular level by the Human Leukocyte Antigen (HLA) family.1
To address this challenge, scientists are exploring methods to overcome HLA-related issues in allogenic iPSC therapies. In this article, we will explore ways to overcome HLA mismatch for allogenic iPSC-derived therapies.
What are Human Leukocyte Antigens (HLA)?
HLAs are the major regulators of the body’s ability to differentiate self from non-self.1 They are the human version of the Major Histocompatibility Complex (MHC) found in all other vertebrates. These surface proteins are found on almost all nucleated cells, and they can be divided into three classes based on their structure and function.
The HLA genes are located on chromosome 6 and are categorized as HLA-A, HLA-B, and so on. HLA class I and class II molecules are crucial for the cellular adaptive immune response as their antigen-presenting role helps immune cells to recognize and respond to foreign molecules perceived as threats.1
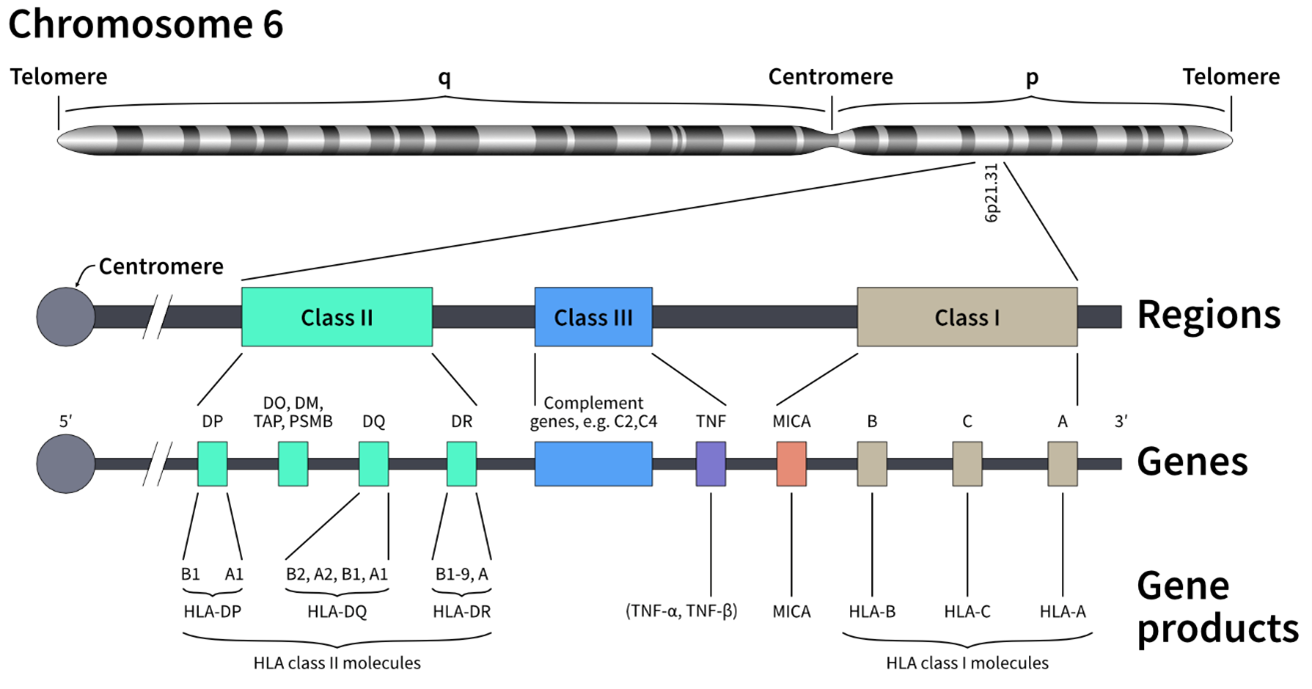
Figure 1: The HLA gene complex is located on multiple locations on chromosome 6 and includes all three classes of HLA molecules.
- The HLA class I gene products (HLA-A, -B, and -C loci) present endogenous peptides mainly from pathogens and tumor cells by forming complexes with β2 microglobulin (B2M) surface proteins, enabling them to regulate interactions with cytotoxic T-cells and trigger cellular immunity.6
- The HLA class II system (HLA-DP, -DQ, and -DR loci) is almost exclusively expressed in antigen-presenting cells (mainly dendritic cells, macrophages and B cells), which process exogenous peptides to present to T cells. On the cell surface they form heterodimers, which subsequently modulates the interaction of T cells with B cells to produce specific antibodies. HLA class II expression is quantitatively controlled by the class II transactivator (CIITA).5
- The HLA class III molecules, in turn, are not adaptable, are part of the innate immune system, and are therefore less important for the creation of hypoimmune cell lines.1
In general, HLA genes show a wide range of genetic variations with more than 35,000 different alleles described.7 Consequently, this diversity (population-specific allele frequencies) could provide an estimation of the chances of finding matched donors in a registry, Allelic diversity is responsible for phenomena such as self vs other recognition and subsequently foreign tissue rejection due to HLA mismatch.1
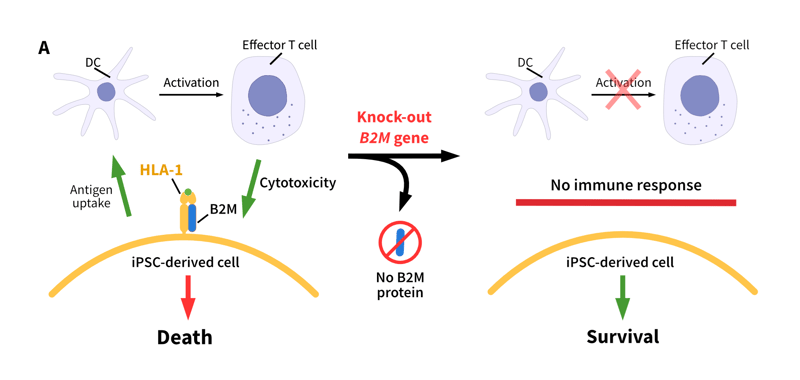
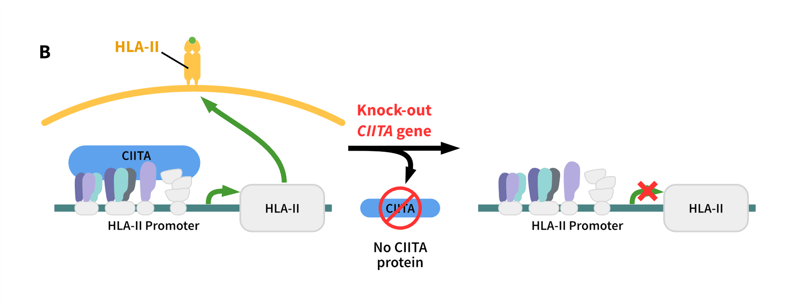
Figure 2: Different strategies to prevent immunogenicity. (A): Knock-out of the B2M gene in human iPSCs deletes the presence of the light chain B2M and disrupts the assembly and presence of HLA-I on the cell surfaces. iPSCs become “unnoticeable” to dendritic cells (DC) and cytotoxic T cells of the host without HLA-I on the surface (B): Knock out of the CIITA gene in human iPSCs directly inhibits the expression of HLA-II and without HLA-II protein complex no HLA-II-dependent immune rejection will be triggered (adapted from Ref 11).
How can we overcome HLA mismatch?
One approach to tackle HLA mismatches when generating cell products for clinical use is to build libraries using numerous iPSC donors with common HLA haplotypes. This strategy is a suitable approach for diseases that show strong linkage to specific HLA alleles, such as narcolepsy, celiac disease, and Type I diabetes.8-10
Yet such a library has a major limitation: The wide range of allelic variation within populations, especially in diverse populations such as the US, would require hundreds of different alleles to account for a significant portion of the population. Each distinct iPSC line in this library would generate a separate therapeutic cell product, requiring independent development processes and approvals.
An alternative approach, which would substantially mitigate the impact of HLAs on self-recognition, involves targeting genes essential for HLA processing machinery such as CIITA and B2M. Knockout of these two genes is one way to achieve this modulation, and several recent studies utilizing CRISPR-Cas9 gene editing have already demonstrated its success.2-4 In the following section we will discuss why it is adequate to eliminate only these two genes to establish a universal iPSC line with hypoimmune properties all while preserving its pluripotency properties.
Examples of CIITA/B2M Knockout
The class II transactivator CIITA is a master regulator for HLA class-II genes and is known to be essential for their transcription: no CIITA expression means no transcription of the HLA Class II system. Therefore, functional loss of the CIITA gene leads to a deficiency in HLA class II molecules, due to the lack of antigen-presentation abilities (Figure 2).2,5 Additionally, by also targeting the B2M gene the immune response can be further suppressed as B2M proteins form a heterodimer with HLA class I proteins, and this complex is therefore required for proper HLA class I presentation on the cell surface. Hence, suppression of B2M expression prevents an immune response from cytotoxic T cells by depleting all HLA class I molecules (Figure 2).2,4,6
Recent studies have shown that knocking out B2M and CIITA in iPSCs can create hypoimmune cells suitable for cell therapeutics. Deuse et al used the gene editing approach to modify both mouse and human iPSCs. They knocked out B2M and CIITA within iPSCs while overexpressing CD47, a ubiquitous cell surface protein that inhibits phagocytosis. This protein supports the evasion of immune rejection and increases long term survival of the engineered cells without the need for supplementation of immunesuppressions.4
In another study by Wang et al, they also turned off expression of B2M and CIITA in human iPSCs, either separately or both together.2 These modified cells were analyzed in monkey models, and the hypoimmunogenic iPSCs showed a decreased immune response, had increased survival, and an enhanced therapeutic effect relative to wild type iPSCs.2 Results for in vitro and in vivo assessments of antigenicity are shown in Figure 3.
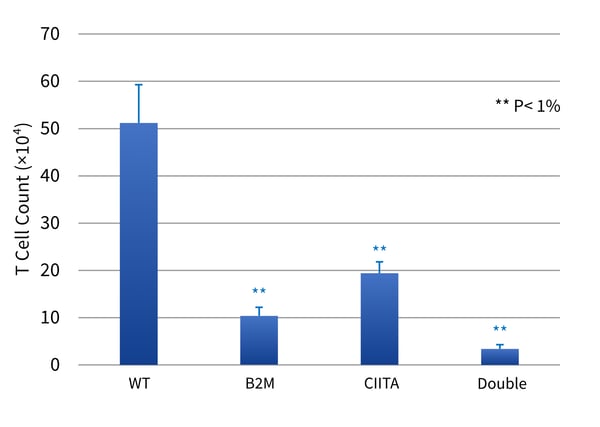
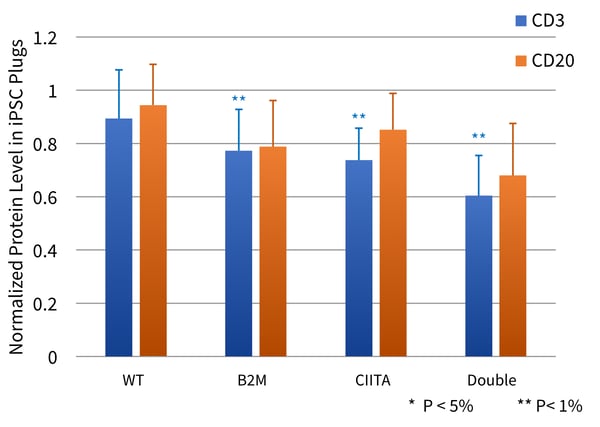
Figure 3 (A; top panel). Human iPSCs (either Wild Type (WT) or single or double mutant, were incubated with allogeneic PBMCs, and the number of T cells were counted by labeling with anti-CD3 and FACS analysis. (B; bottom panel) WT or single or double mutant iPSCs were imbedded in Corning® Matrigel® and implanted subcutaneously in monkeys. After 3 days, the accumulation of T cells (blue bars; CD3+) and B cells (orange bars; CD20+) was estimated by quantitative Western blotting. P values relative to WT (T test) are indicated by * and **.
Outsourcing clinical gene editing of iPSCs
If you are looking to outsource your gene editing projects, REPROCELL provides a StemEdit Clinical Gene Editing Service. This service employs the power of our CRISPR technology to make specific changes to important genes like B2M and CIITA in clinically relevant iPSCs (Figure 2).
For more information about this service, please check our website or reach out to us via email info-us@reprocell.com.
References
- Klein and Sato. The HLA System. N Engl J Med 343:702 (2000).
- Wang X et al. Diminished expression of major histocompatibility complex facilitates the use of human induced pluripotent stem cells in monkey. Stem Cell Research & Therapy 11:334 (2020).
- Xu H et al. Targeted Disruption of HLA Genes via CRISPR-Cas9 Generates iPSCs with Enhanced Immune Compatibility. Cell Stem Cell 24 :566 (2019).
- Deuse et al. Hypoimmunogenic derivatives of induced pluripotent stem cells evade immune rejection in fully immunocompetent allogeneic recipients. Nature Biotechnology 37:252 (2019).
- Steimle V et al. Regulation of MHC class II expression by interferon-γ mediated by the transactivator gene CIITA. Science 265:106 (1994).
- Lu P et al. Generation hypoimmunogenic human embryonic stem cells by the disruption of beta 2-microglobin. Stem Cell Rev Reports 9:806 (2013).
- EMBL-EBI. Welcome to IPD-IMGT/HLA Release 3.15 (2023).
- Chabas et al. The genetics of narcolepsy. Annu Rev Genomics Hum Genet 4:459 (2003).
- Noble and Valdes. Genetics of the HLA region in the prediction of Type 1 Diabetes. Curr Diab Rep 11:533 (2011)
- Sciurti et al. Genetic susceptibility and celiac disease: what role do HLA haplotypes play? Acta Biomed 89 Suppl 9: 17 (2018).
- Zhang et al. Concise review: One stone for multiple birds: Generating universally compatible human embryonic stem cells. Stem Cells 34:2269 (2016).