Extraction of RNA from Cells Cultured in Alvetex® Scaffold in 96-Well Plate Format
● Download this protocol as a PDF (0.2 MB)
1. Introduction
This document describes two methods for the extraction of RNA from cells cultured in the Alvetex Scaffold 96 well plate (AVP009). RNA extraction can be performed in situ without prior recovery of cells. Both methods use commercial RNA extraction reagents/kits.
2. Methods
Two methods are described for RNA isolation, using the following commercially available products:
- TRIzol® Reagent (Invitrogen (Thermo Fisher), 15596026)
- RNeasy® Plus Universal Mini Kit (Qiagen, 73404)
2.1. Method 1: TRIzol® Reagent
This is an example protocol, based on 3D culture of MDA-MB-231 breast cancer cells in Alvetex Scaffold in 96-well plates. RNA was extracted using the TRIzol® reagent (Invitrogen (Thermo Fisher), 15596026).
- Aspirate culture medium and wash cells with 200 μL PBS, aspirate.
- Add 200 µL TRIzol® reagent and place on shaker for 10 minutes, 100 rpm (room temperature).
- Lyse cells by pipetting up and down × 10 and transfer lysate to microcentrifuge tube.
- Add 80 μL chloroform and vortex for 15 seconds.
- Centrifuge at 12000 × g for 15 minutes at 4 °C.
Note: RNA remains in upper aqueous phase.
- Remove upper aqueous phase to a clean tube (do not collect any interphase or organic phase material).
- Add 30 μg glycogen (GlycoBlue, Invitrogen AM9515) as co-precipitator.
- Add 200 µL 100 % isopropanol.
- Incubate at room temperature for 10 minutes.
- Centrifuge at 12000 × g for 10 minutes.
- Carefully discard supernatant to leave RNA pellet in the tube.
- Wash RNA pellet with 200 µL 75% ethanol.
Note: sample can be stored in this state for 1 year plus at –20 °C, or 1 week at 4 °C.
- Briefly vortex and centrifuge at 7500 × g for 5 minutes at 4°C. Discard the wash supernatant.
- Air-dry the RNA pellet for 5-10 minutes.
Note: do not allow pellet to dry completely.
- Resuspend pellet in 50 µL RNase-free water by pipetting up and down.
- 16. Incubate in a water bath or heat block at 55-60°C for 10-15 minutes.
- Store sample at –80°C.
2.2. Method 2: RNeasy® Plus Universal Mini Kit
This is an example protocol, based on 3D culture of MDA-MB-231 breast cancer cells in Alvetex Scaffold in 96-well plates. RNA was extracted using the RNeasy® Plus Universal Mini Kit (Qiagen, 73404).
Before starting:
- Add 2 volumes of ethanol to buffer RWT as indicated on bottle, to achieve working dilution.
- Add 4 volumes of ethanol to buffer RPE as indicated on bottle, to achieve working dilution.
- Aspirate medium and wash cells with 200 µL PBS, aspirate.
- Add 180 µL QIAzol Lysis Reagent to each well and place on a shaker (100 rpm) for 10 minutes.
- Homogenise samples by pipetting up and down × 15 and transfer lysates to clean micro-centrifuge tubes, incubate at room temperature for 5 minutes.
- Add 20 µL gDNA Eliminator Solution and vortex for 15 seconds.
- Add 40 µL chloroform and vortex for 15 seconds.
- Place upright and incubate at room temperature for 2-3 minutes.
- Centrifuge at 12 000 × g for 15 minutes at 4 °C
Note: RNA will partition into the upper aqueous phase, approx 120 μL in total.
- Transfer upper aqueous phase to clean tube.
- Add 1 volume of 70 % ethanol and pipette up and down to mix.
- Transfer sample to an RNeasy® spin column placed in a 2 mL collection tube. Close lid and
centrifuge at ≥ 8000 × g for 15 seconds at room temperature. Discard flow-through. - Add 700 μL Buffer RWT to the RNeasy® spin column. Close lid and centrifuge at ≥ 8000 × g for
15 seconds at room temperature, to wash the membrane. Discard flow-through. - Add 500 μL Buffer RPE to the RNeasy® spin column. Close lid and centrifuge at ≥ 8000 × g for
15 seconds at room temperature, to wash the membrane. Discard flow-through. - Add 500 μL Buffer RPE to the RNeasy® spin column. Close lid and centrifuge at ≥ 8000 × g for2
minutes at room temperature, to wash the membrane. - Place the RNeasy® spin column in a clean 2 mL collection tube and centrifuge at full speed for
1 minute. - Place the RNeasy® spin column in a clean 1.5 mL collection tube and add 50 μL RNase-free
water directly to the spin column membrane. Close lid and centrifuge at ≥ 8000 × g for
1 minute. - Repeat step 16 either (i.) by passing the first flow-through eluate through the column again,
or (ii.) using a second volume of RNase-free water
Note: (i.) will result in a 15-30% lower RNA yield, while (ii.) will yield a more dilute RNA sample.
3. Example Data
A
| ng/mL | SD | µg/well | |
|---|---|---|---|
| 2D | 59.8 | 10.5 | 10.5 |
| Alvetex Scaffold | 63.3 | 14.3 | 3.17 |
B
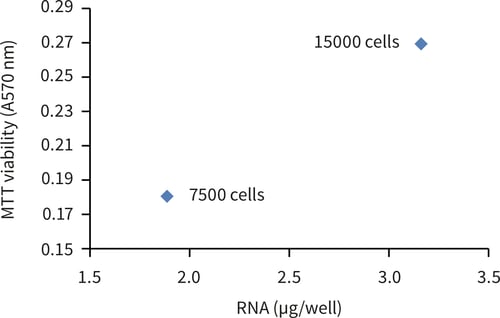
Figure 1. RNA extraction from Alvetex Scaffold 96-well plate using TRIzol® reagent. RNA was extracted from cultures of MDA-MB-231 cells cultured for 24 hours. (A.) Comparable RNA yield were obtained from standard 2D and Alvetex Scaffold 96-well plates. (B.) Quantitative increase in extracted RNA between 7500 and 15000 cells per well in Alvetex Scaffold 96-well plates. Data represent mean of n = 2.
B

Figure 2. RNA extraction from Alvetex Scaffold 96-well plate using RNeasy® Plus Universal Mini Kit. RNA was extracted from cultures of MDA-MB-231 cells seeded at 20,000 cells per well and grown for 24 hours. (A.) Comparable RNA yields were obtained from standard 2D and Alvetex Scaffold 96-well plates. Data represent mean of n = 2. (B.) Spectrophotometric traces of RNA samples extracted from MDA-MB-231 cultures in standard 2D and Alvetex Scaffold 96-well plates. Spectra are typical ofdilute RNA samples obtained using RNeasy® reagents; RNA absorbance at 260 nm overlaps with phenol absorbance at 270 nm (residual QIAzol Lysis Reagent), while residual guanidine isothiocyanate absorbs strongly below 230 nm (residual QIAzol Lysis Reagent and Buffer RWT).