Live Cell Imaging of 3D Cultures Grown on Alvetex® Scaffold Using Confocal Microscopy
● Download this protocol as a PDF (2.4 MB)
1. Introduction
Live cell imaging allows real time monitoring of cell shape and form during cell differentiation, proliferation and migration. Alvetex Scaffold offers a unique ability to study cell growth in 3D by providing an environment through which individual cells can navigate for extended periods of time and where confluent cell populations routinely multilayer. Below are example results for live cell labelling of 3D cultures in Alvetex Scaffold. It is recommended that for each staining agent used, the original manufacturers’ instructions be followed. Imaging of the specimens will also need to be optimised according to the specifications of the microscope used.

Figure 1: CHO-K1 cultures grown in 6 well inserts were transferred to a 35mm plastic cell culture dish (Nunc, TKT100030M) and placed on a heated stage insert fitted to a ZEISS LSM 510 confocal microscope with live cell imaging capability. The culture was maintained in HEPES-buffered F-10 medium during imaging.
2. General Recommendations for Live Cell Imaging
Successful live cell imaging requires the careful optimisation of experimental conditions and microscope set-ups. In addition to following manufacturer’s instructions, below are a few examples of simple parameters that can greatly affect the quality of the results obtained. These are even more important for long exposures, i.e. experiments ranging from overnight to several days.
Stage inserts designed to maintain the cell culture plate at a temperature of 37°C require overnight equilibration to allow for the expansion of the metal frame. Insufficient equilibration times will not result in full expansion of the frame and lead to a continuous loss of focus until equilibrium is reached, i.e. up to several hours.
CO2 supply designed to maintain a suitable pH within the cultures requires a few hours runtime prior to introduction of the cultures to the containment cage. In cases when the CO2 supply is absent or unstable, HEPES-buffered cell culture media can be used.
Repeated and/or long-term exposure of dyes and live cells to fluorescent light can lead to photobleaching and light toxicity, respectively. These problems can be reduced by optimising laser settings and making sure that shutters are closed in between image acquisitions.
3. Recommendations Specific to Cultures Grown in Alvetex Scaffold
It is widely appreciated that visualisation of cells inside intact tissues is limited using conventional microscopy methods. Similarly, when viewing an unstained, unsectioned Alvetex Scaffold 3D culture under a standard light microscope, the combined density and thickness of the scaffold and the 3D culture within it prevent the clear visualisation of individual cells. Therefore, cells grown in Alvetex Scaffold must be stained prior to imaging and, whenever possible, best results will be obtained by using fluorescent probes combined with confocal microscopy.
Alvetex Scaffold discs are supplied at a thickness of 200 μm, which might exceed the Z-stack capacity of some models of confocal microscopes. The exact depth of sample that can be successfully imaged will vary according to the density of the cell culture and the fluorophore used, so that depths of between 50 and 100 μm can be achieved from a cell-seeded Alvetex Scaffold disc. As high-density cell cultures grown in Alvetex Scaffold approximate the complexity and structure of in vivo tissues, fluorophores specifically developed for in vivo deep imaging can be used to improve performance if needed.
The material used to manufacture Alvetex Scaffold will produce minimal autofluorescence and will not result in background signal when using confocal microscopy. However, any hydrophobic dye (e.g. DiI, calcein) will bind strongly to Alvetex Scaffold and must therefore be added to the cells prior to seeding into Alvetex Scaffold.
Correct focus will only be achieved within the working range of the objective. Therefore if using Alvetex Scaffold in an insert format, it might be necessary to lift the insert from its holder and to place it at the bottom of a Petri dish filled with the appropriate cell culture medium. Then image the 3D culture in the Petri dish and return the Alvetex Scaffold disc to its original holder afterwards if applicable.
4. Materials
4.1. General Materials
- Cells: e.g. CHO-K1 cells (ATCC, CCL-61).
- Imaging media: e.g. for CHO-K1: Ham’s F-10 nutrient mixture, HEPES-buffered (Gibco, Catalogue No: 22390025), supplemented with 10% (v/v) foetal calf serum, 100 µg/mL penicillin and 10 µg/mL streptomycin. Note: Gibco’s F-10 already contains L-glutamine.
- Alvetex Scaffold in 6 or 12 well inserts (AVP004/AVP005).
4.2. Materials Required for Fluorescent Labelling
GFP Labelling:
- Transfection reagent: e.g. linear, 25 kDa polyethylenimine (PEI) (Park Scientific Ltd., Catalogue No: 23966-2), diluted to a stock concentration of 0.9 mg/mL in dimethylsulfoxide (DMSO) (Sigma-Aldrich, Catalogue No: D-8779), followed by filtration through a 0.2 μm mesh, aliquotted and stored at –20 °C.
- Vector DNA: e.g. gWIZ GFP mammalian expression vector (Amsbio, Catalogue No: P040400), provided at working concentration (1µg/µL) and stored at –20 °C.
DiI Labelling:
- CellTracker™ CM-DiI (Invitrogen, Catalogue No: C-7000), diluted to a stock concentration of 2 mg/mL in dimethylformamide (DMF) (Sigma-Aldrich, Catalogue No: D-4551) and stored at –20 °C.
Hoechst Counterstaining:
- Hoechst 33342 (Invitrogen, Catalogue No: H-3570), as supplied at the stock concentration of 10 mg/mL and stored at 4°C.
4.3. Equipment and Materials Required for Imaging
Confocal fluorescent microscope with live cell imaging capacity, i.e. time-lapse acquisition, temperature-controlled stage and CO2 supply. HEPES-buffered cell culture media can be used in cases where a CO2 supply is unavailable. In the example results below, a ZEISS LSM 510 was used.
5. Methods
5.1. DiI Labelling
Note: Hydrophobic dyes (eg. DiI, calcein) can be used to label cells grown on Alvetex Scaffold, but the dye must be added prior to cell seeding and the cells thoroughly washed after staining to minimise binding of residual dye to Alvetex Scaffold. As some hydrophobic dyes will partition between dividing cells, the time in culture before imaging must also be kept in mind to prevent excessive dye dilution within a growing cell population.
- Expand populations of cells in culture medium as monolayer cultures in conventional 2D plastic-ware according to standard procedures.
- See further information at Alvetex Protocols on how to choose, handle and prepare the Alvetex Scaffold for cell culture.
- Prepare a single-cell suspension of cells in media using trypsin-EDTA to remove cells from 2D plastic-ware. Count cells and prepare seeding suspensions in medium to allow for a seeding density of 1 × 106 cells in 3.5 mL, per well.
- Add CellTracker™ CM-DiI at a dilution of 1/1000 of the stock solution. Mix and keep at 37 °C for 5 minutes. Protect from light.
- Spin down for 5 minutes at 1000rpm. Remove supernatant and resuspend in 3.5 mL of appropriate cell culture medium. Spin down again for 5 minutes at 1000 rpm. Remove supernatant and resuspend to allow for a seeding density compatible with the Alvetex Scaffold format used.
5.2. GFP Labelling
Note: Although this example protocol describes the transfection of live cells in suspension before seeding into Alvetex Scaffold, adherent 3D Alvetex Scaffold cultures can also be transfected and imaged.
- Expand populations of cells in culture medium as monolayer cultures in conventional 2D plastic-ware according to standard procedures.
- See further information at Alvetex Protocols on how to choose, handle and prepare the Alvetex Scaffold for cell culture.
- In a sterile 1.5 mL tube, mix 2 μL of vector DNA with 11.11 µL of PEI solution, i.e. equivalent to a DNA:PEI ratio of 1:5 (w/w), per well. Leave for 15 minutes at room temperature.
- Meanwhile, prepare a single-cell suspension of cells in media using Trypsin-EDTA to remove cells from 2D plastic-ware. Count cells and prepare seeding suspensions in medium to allow for a seeding density of 1 × 106 cells in 0.5 mL, per well.
- Add mL of cell suspension to the DNA:PEI solution and mix gently. This is equivalent to 2 µg DNA per million cells.
- Seed cells on Alvetex Scaffold according to the format used and leave the cultures in a 37 °C, 5 % CO2 cell culture incubator. Replace the medium with appropriate fresh cell culture medium the following day and return the cultures to the 37 °C, 5 % CO2 cell culture incubator until the desired imaging timepoint. Protect from light.
5.3. Hoechst 33342 Counterstaining
Note: For all the example labelling protocols described above (sections 1.0 and 2.0), cells can be counterstained with Hoechst 33342 to check for labelling efficiency. Shortly prior to imaging, add Hoechst 33342 at a dilution of 1/1000 of the supplied stock solution. Leave for 30 minutes at room temperature, then replace the medium with fresh cell culture medium and proceed with imaging. Protect from light until imaging.
6. Example Results
CHO-K1 cells (ATCC, CCL-61) were maintained in F-12K medium (Kaighn’s modification, Gibco, 21127022) supplemented with 10 % (v/v) foetal calf serum, 100 μg/mL penicillin and 10 μg/mL streptomycin (for full methods refer to Example protocol for the culture of CHO-K1 cells on Alvetex Scaffold located at reinnervate.com/using-alvetex/protocols. For imaging, cultures were switched to Ham’s F-10 nutrient mixture buffered with HEPES (Gibco, 22390025), supplemented with 10 % (v/v) foetal calf serum, 100 µg/mL penicillin and 10 μg/mL streptomycin.
Below are captured images of live cells over a time-course: labelled with CellTracker™ (Figure 2.) and cells transfected with GFP (Figure 3.).
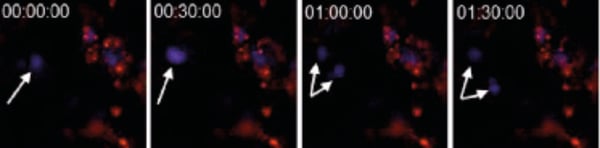
Figure 2. CHO-K1 cells were stained with CellTrackerTM CM-DiI (1/1000 dilution) immediately prior to seeding onto Alvetex Scaffold 6-well inserts. After five days, cultures were imaged on a ZEISS LSM 510 confocal microscope. Cells were stained with Hoechst 33342 the evening prior to imaging. Note the dividing cell (arrows).
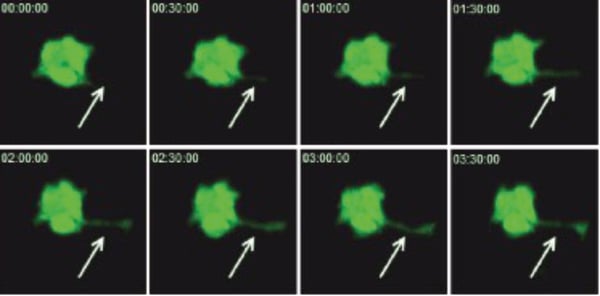
Figure 3. CHO-K1 cells were grown on Alvetex Scaffold 6-well inserts. After five days, cultures were transfected with G-Wiz GFP construct and left to recover for a further 24 hours before imaging on a ZEISS LSM 510 confocal microscope. Pictures shown are flattened Z-stacks (6 sections, total height 18 µm). Note the development of a cytoplasmic extension during the 3:30 hour recording period (arrows).