Protein Extraction from Cells Cultured in Alvetex® Scaffold 96-Well Plate Format
● Download this protocol as a PDF (0.3 MB)
1. Introduction
This protocol describes the use of a commercial protein extraction reagent to obtain protein samples from cells cultured in Alvetex Scaffold 96-well plate format (AVP009). Using this method, quantitative protein extraction is achievable over wide ranging cell densities. Resulting protein extracts are compatible with common downstream techniques, including Bradford and BCA protein assays, SDS-PAGE and western blotting, non-denatured immunological applications (e.g. immunoprecipitation), and enzymatic assays.
2. Method
This is an example protocol, based on 3D culture of MDA-MB-231 breast cancer cells in Alvetex Scaffold in 96-well plates. Total protein was extracted using the Mammalian Protein Extraction Reagent (M-PER) from Pierce (now Thermo Fisher) (product code 78503).
- Withdraw culture medium from each well of the Alvetex Scaffold 96-well plate.
- Rinse cells by carefully adding 200 μL PBS per well, aspirate to waste.
- Add 100 μL M-PER reagent to each well and place plate on a shaking platform (100 rpm for
10 minutes).
Note: The M-PER reagent can be supplemented with protease inhibitors (e.g. Pierce, 87785) to minimise protein degradation upon release of cellular proteases.
- Agitate the liquid in each well by pipetting up and down, and transfer to 0.5 mL microcentrifuge tubes.
- Centrifuge samples at 14,000 × g for 5 minutes to remove cell debris.
- Transfer supernatants to clean tubes and store at –80 °C.
3. Example Data
MDA-MB-231 breast carcinoma cells (ATCC, HTB-26) were routinely maintained in T-75 flasks. MDA-MB-231 culture media consisted of: Leibovitz’s L-15 medium (ATCC, 30-2008) supplemented with 10 % v/v FBS and 100 U/mL Penicillin/ Streptomycin. Cells were seeded into Alvetex Scaffold 96-well plates (AVP009) at seeding densities ranging from 25 × 103 to 200 × 103 cells in a total media volume of 200 μL per well. Plates were incubated for 24 hours at 37 °C with 0 % CO2, after which cultures were processed according to the protocol above.
3.1. Protein analysis results
Determination of relative protein concentration was made using Protein Assay Reagent (Bio-Rad, 5000006). A dilution series of BSA was used for the generation of a standard curve (Figure 1). Approximately 1 milligram of protein was isolated per disc (Table 1).
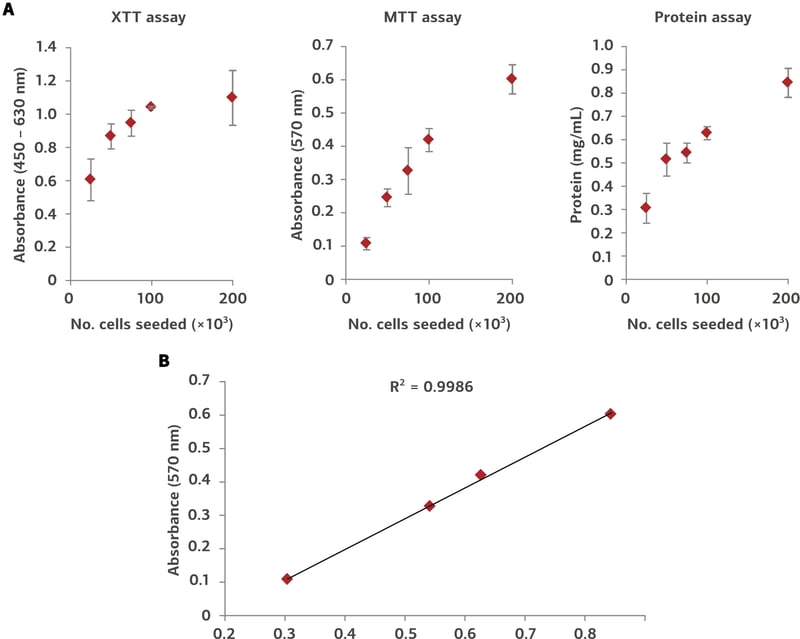
Figure 1. Protein extraction from Alvetex Scaffold 96-well plate. MDA-MB-231 cells were seeded into Alvetex Scaffold 96-well plates at densities of 25, 50, 75, 100, and 200 × 103 cells in 200 µL per well. Cell viability was measured using two methods (XTT and MTT assays) for comparison with protein concentrations in extracts obtained via the protocol above (A) (n = 3, mean ± SD). Pairwise plot of MTT viability and protein concentration (B) reveals a linear relationship between cell number and protein yield (R2 = 0.9986). Protein yields ranged from approximately 30 µg/well for 25 × 103 cells seeded, to approximately 85 µg/well for 200 × 103 cells.
| No. cells seeded per well | Protein yield per 3D culture (µg/mL) |
|---|---|
| 25,000 | 30.5 |
| 50,000 | 51.4 |
| 75,000 | 54.3 |
| 100,000 | 62.8 |
| 200,000 | 84.4 |
Table 1. Example yields of protein extraction from Alvetex Scaffold 96-well plate. Protein was estimated using the Protein Assay Reagent (Bio-Rad, 5000006) and a BSA standard curve. For protocols for XTT and MTT viability assays in Alvetex Scaffold 96-well plates see: Alvetex Protocols. Approximate protein yields in extracts from Alvetex Scaffold cultures of HepG2 hepatocytes, using BSA as a standard reference. Values shown represent the mean of triplicate readings per sample (±SD).