Alvetex Scaffold Application Note 15
3D cell culture: mimicking real tissue: Running Alvetex® Scaffold in 96-well plate format on the TECAN Freedom EVO®
Download this application note as a PDF (2.04 MB)
Introduction
Experiments performed in a classical two-dimensional (2D) cell culture system have resulted in a large body of knowledge about basic life science. This type of research began in the 1880s, and has provided numerous answers about intrinsic cellular biology and develop-mental molecular processes, as well as enabling break-throughs in regenerative medicine and insights into diseases and drug discovery. However, the 2D cell culture systems used so far have several drawbacks: the morphology, proliferation, metabolism and expression profiles of cells grown in 2D systems are very different to cells in living tissues.
Technological advances in the last couple of years have enabled 3D scaffolds to be engineered that offer a real proxy for the in vivo environment which can be easily and routinely used in a cell culture laboratory. The Alvetex Scaffold (Figure 1) is a 200 μm thick, highly porous and uniform polystyrene scaffold, designed and developed by REPROCELL Europe (Sedgefield, UK)[1].
Alvetex Scaffold enables cells to develop natural, in vivo-like intercellular interactions, providing an ideal environment for real three-dimensional growth and nutrient exchange that is comparable to intra-capillary exchange in living tissues.
This study demonstrates that the TECAN Freedom EVO work-station can simply and routinely process cell cultures in Alvetex Scaffold in a 96-well plate format. Two independent assays were performed to prove the quality of the culture.
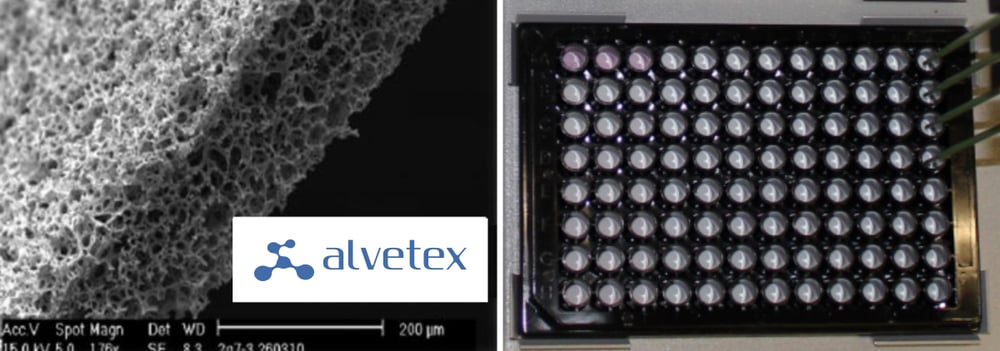
Figure 1. Left: SEM of the Alvetex Scaffold made from polystyrene. Average void size 40μm. Right: photograph of an Alvetex Scaffold 96-well plate in SBS format with four fixed tips from a Freedom EVO Liquid Handling (LiHa) Arm.
Materials and Methods
Liquid handling equipment
The Freedom EVO platform used in this study is equipped with a Liquid Handling (LiHa) Arm with eight fixed tips, a Robotic Manipulator (RoMa) Arm and an orbital shaker (Figure 2). Other modules shown in Figure 2 were not used in the protocols described. The workstation was housed in a biological safety cabinet, and experiments were conducted under sterile conditions.
Experimental conditions
Alvetex Scaffold in 96-well plate format is composed of a black plate with a clear plastic base. To render the scaffold hydrophilic, each well was preconditioned with 300 μl of 70 % ethanol. Every well was subsequently washed twice with 300 μl of PBS, followed by two washes with growth medium (RPMI-1640: Sigma R8758, supplemented with 10 % FCS, 1 % Pen/Strep). HCT-116 (colon cancer cell line) cells, kindly provided by Zurich University of Applied Sciences (ZHAW, Wädenswil), were seeded at a density of 10,000 cells per well in 200 μl of growth medium, and cultivated at 37 °C with 5 % CO2. Cells were cultured for eight days and each experiment was performed in duplicate. The medium was exchanged every two days during the first five days of incubation, and daily thereafter.
Cell viability was tested using the CellTiter-Glo® Luminescent Cell Viability Assay, (Promega, G7571) and DNA quantification was performed with the Quant-iT™ PicoGreen® dsDNA Assay Kit (Invitrogen, P11496). After cultivating the cells for two, five or eight days, results were analysed using a multimode plate reader (BMG Labtech, FLUOstar OPTIMA). Prior to performing the assays, cell lysis was carried out using the lysis buffer included in the Promega CellTiter-Glo kit.
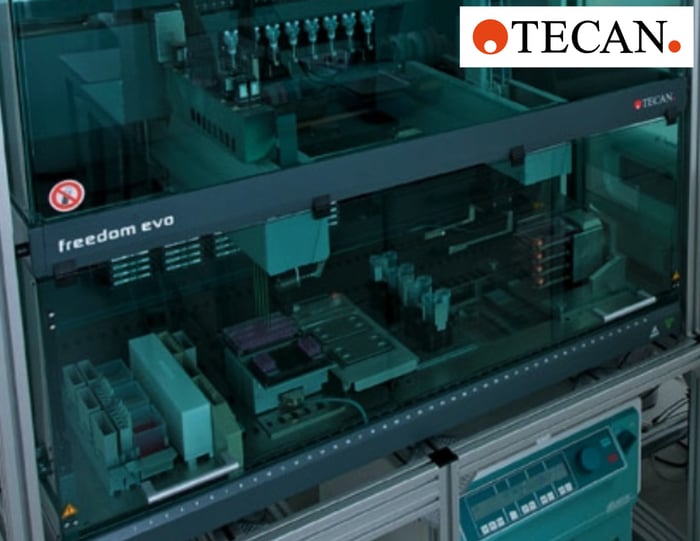
Figure 2. The Freedom EVO cell biology workstation (TECAN).
Automated CellTiter-Glo Luminescent Cell Viability Assay
The CellTiter-Glo Assay is used to determine the number of viable cells in a culture based on quantification of the amount of ATP, which indicates the presence of metabolically active cells. A ‘glow-type’ luminescent signal is produced by a luciferase reaction, and is directly proportional to the number of cells present in the culture. After two, five and eight days in the culture, the medium was removed from the respective wells and replaced with 75 μl of fresh medium and 75 μl of Cell Titer-Glo Reagent. After incubation at room temperature for two minutes while shaking (1000 rpm), then an additional 10 minute incubation without shaking followed by a further one minute shaking period, 50 μl of the reaction solution was aspirated and dispensed into half area, white 96-well plates (Greiner Bio-One, Ref.: 675075) for analysis. Luminescence measurements were performed in a multi-mode plate reader.
Manual DNA quantification
The DNA quantification assay is based on PicoGreen, a dye that labels double-stranded DNA (dsDNA) in solution. A further aliquot was taken from the luciferase reaction described above, aspirating 50 μl from the respective wells after two, five and eight days in culture.
The aliquot was transferred to a separate black 96-well plate (Greiner Bio-One, Ref.: 655076) containing 50 μl of PBS. After addition of 100 μl of PicoGreen solution (1:200 in 1× TE buffer) and incubation at room temperature for five minutes, fluorescence emission intensity was measured on a multimode reader (485 nm Ex, 520 nm Em). Figure 3 shows a comparison of results from two, five and eight days in culture.
Results
The cell viability assay shows a typical growth curve for the HCT- 116 cells over time (Figure 3). These data are in line with results obtained from cells cultivated in dextran hydrogels (unpublished data, Rimann et al.). The initial logarithmic growth during the first five days, corresponding to increased cell proliferation, is followed by less intense growth from day five to day eight, with a tendency to plateau.
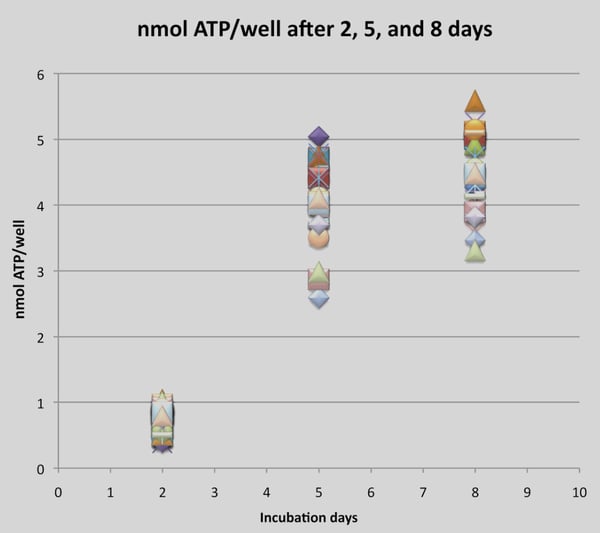
Figure 3. Cell viability assay (CellTiter-Glo assay) of HCT-116 cells. Each data point represents the measured concentration of ATP in a single well after two, five and eight days in 3D culture.
Identification of dsDNA in the supernatant of the Alvetex Scaffold 3D cultures using the Quant-iT PicoGreen assay showed that the amount of dsDNA significantly increases over time (Figure 4).
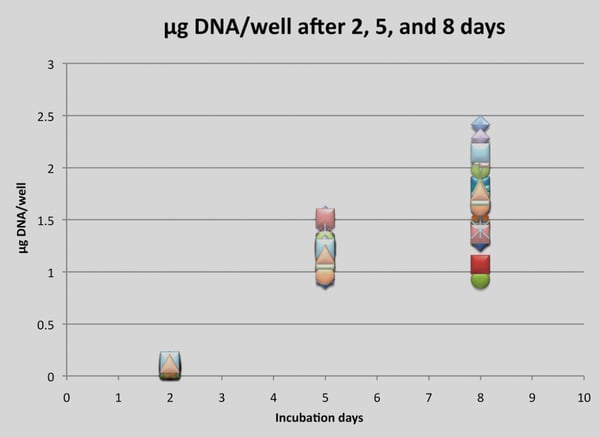
Figure 4. DNA quantification (Quant-iT PicoGreen Assay Kit) of HCT-116 cells. Each data point represents a single DNA concentration of a well after two, five and eight days in 3D culture.
Discussion
Enabling automation of Alvetex 3D cell culture on the Freedom EVO workstation
The cells cultivated on the Freedom EVO platform in Alvetex Scaffold 96-well plates propagate and show viability comparable to growth in manual 3D culture using established Alvetex Scaffold protocols (see online technical support at Protocols). Tedious and redundant manual approaches are replaced with optimized protocols for automation on a Freedom EVO workstation. This reliable and robust process reduces the variability that is potentially introduced by human intervention, and therefore offers more biologically relevant data.
Expansion of HCT-116 cells on Alvetex Scaffold 96-well plates
The CellTiter-Glo Luminescent Cell Viability Assay and Quant-iT PicoGreen dsDNA Assay Kit are classical viability assays that show growth and proliferation of cells in culture. These assays are standard methods in a variety of research areas, such as toxicity testing, developmental biology research and cancer research, as well as early cell-based screens in target identification and validation of drug discovery.
The cell viability assay clearly indicates the number of proliferating and therefore healthy cells. Quantification of the double-stranded DNA gives only a rough estimate of all cell material, including cell debris and dead cells. However, this is still a very valuable tool for verification of the data from the cell viability assay.
Both assays show that cells in Alvetex Scaffold 96-well plates handled on an automated Freedom EVO work-station grow and proliferate in a 3D cell culture environment. The results are comparable to manual cell culture using Alvetex, and support the idea that cells grow and propagate better in cell cultures mimicking tissue-like architecture.
Conclusion
Alvetex Scaffold provides an in vivo proxy for performing a variety of cellular and molecular techniques and bio-chemical assays, resulting in more biologically relevant data. When combined with the Freedom EVO liquid handling platform, cell biologists will be able to achieve a higher throughput, as well as a higher level of standardization for both basic research and drug discovery.
Acknowledgements
The experiments were performed on a Freedom EVO liquid handling platform at Zurich University of Applied Sciences, Institute of Chemistry and Biological Chemistry, Wädenswil. We are grateful to Markus Rimann for performing the experiments and analyzing the data.