Alvetex Scaffold Application Note 04
Preclinical Profiling of Anti-Cancer Therapeutics Using a Novel 96-Well 3D Cell Culture Assay
Download this application note as a PDF (1.2 MB)
Dr Sumeer Dhar
Oncotest GmbH – Institute for Experimental Oncology, Am Flughafen 1 2-1 4, 791 08 Freiburg, Germany.
Professor Stefan Przyborski
REPROCELL Europe Ltd. Glasgow, UK.
Durham University, UK.
stefan.przyborski@reprocell.com
Abstract
Three-dimensional (3D) tumor cell culture models have been shown to represent a biologically relevant assay system, feasible for the development of robust preclinical anti-cancer drug screening platforms. The major advantages of cells cultured in a 3D format are that these retain in vivo-like structures and cell signalling patterns. The influence of non-tumor cells and elements of the tumor microenvironment can modelled using co-culture approaches. Oncotest and REPROCELL (formerly Reinnervate*) have developed agar-free 3D cell culture formats using Alvetex Scaffold for preclinical ex vivo drug efficacy tests to fill the gap between the existing two-dimensional (2D) culture methods and preclinical in vivo model systems Alvetex Scaffold enables a straight-forward and reproducible 3D culture of both tumor cell lines and primary tumor cells, e.g. single cell suspensions prepared from patient-derived xenografts. The new 96-well Alvetex Scaffold format is compatible with automated medium to high throughput drug activity screening. Here we report the development of assays to assess anti-cancer drug activity using the Alvetex Scaffold 96-well plate. A major advantage of the agar-free 3D cell culture is the simple retrieval of cellular material post-drug treatment for subsequent molecular analyses and mechanism of action studies.
*Reinnervate Ltd was acquired by REPROCELL Inc in 2014. It was merged with Biopta Ltd to form REPROCELL Europe Ltd.
Introduction
The screening and assessment of new chemical entities and biologics, to test for anti-tumor activity, has for many years been carried out on conventional two-dimensional (2D) monolayer cell cultures in 96- and 384-well plates. These assays are simple to set up, can be automated and take advantage of luminescent assay read-outs for the reproducible generation of large data sets. Although monolayer cell culture showed initial promise in the discovery of drugs for cancer therapy, many of these potentially active drugs do not perform as well in preclinical in vivo model systems and subsequently display a disappointing success rate in the clinic. One major reason for the poor predictive power of these assays is that cells cultured in monolayer possess an abnormal flattened structure that compromises diverse cellular functions.
Over the past two decades, soft-agar based 3D cell culture assays (clonogenic assay) have been used successfully to assess the drug response of non-adherent cancer cells in semi-solid supports, thereby proving to be a valuable alternative to the routine 2D cell culture. However, there are also certain process-related limitations to the existing clonogenic soft agar growth assays, especially regarding throughput and recovery of biological material post drug treatment. With the development of Alvetex Scaffold technology, there is the opportunity to significantly advance current practice by overcoming existing practical issues, creating physiologically relevant cell-based assays, and enabling efficient and reproducible assessment of cancer cell cytotoxicity. Alvetex Scaffold is a market leading product developed by REPROCELL and is designed to enable researchers to culture cells routinely in 3D. Within the 3D environment of Alvetex Scaffold, cells retain their natural 3D architecture and do not form flattened structures as in conventional 2D cultures. This retention of normal 3D structure provides a more physiologically relevant model, where adjacent cells can grow together in 3D, forming tissue-like structures. In this article we describe the application of the Alvetex Scaffold 96-well plate in the development of 3D culture assays to investigate the toxicity of anticancer agents on various types of human tumor cells.
This study was conducted in close collaboration with Oncotest, a contract research organisation providing pre-clinical profiling services to pharmaceutical companies and academic research institutes developing novel anti-cancer therapies. Oncotest has more than 20 years of experience using both 3D cell culture and xenograft technology to overcome the limitations posed to the oncology drug development process by 2D monolayer cultures. Oncotest’s unique collection comprises over 300 well characterized patient-derived xenograft tumor models (PDX) and more than 60 proprietary cell lines established from patient-derived xenograft tumor models in-house. The PDX-based assay system includes in vitro, ex vivo and in vivo approaches, enabling pharmaceutical partners to develop more effective therapies and screen out ineffective anti-cancer molecules earlier in the drug development cycle.
As more and more targeted drugs are being developed, the need for the assessment of transcriptional and post-translational effects of novel anti-cancer agents is becoming highly pertinent. These investigations are challenging when using an agar-based clonogenic assay. Therefore, Oncotest has been exploring new methodological approaches to study target-related effects and downstream signalling pathways post drug treatment in a physiologically relevant 3D assay.
After careful evaluation of several technologies, Oncotest have adopted Alvetex Scaffold 96-well 3D culture plates because they found them the simplest and most reliable technology on the market today. In this article, we demonstrate the application of the new Alvetex Scaffold 96-well 3D culture plates for the growth and assessment of human cancer cells in response to anti-cancer agents in an automated medium throughput setting. Specifically, we show:
- Robust and reproducible 3D culture of human cancer cells;
- Assessment of cytotoxicity using Promega’s CellTiter-Glo® luminescent cell viability assay;
- Assessment of monotherapies and combined drug treatments in 3D culture;
- Compatibility with automated work stations such as Tecan’s EVO 200 platform;
- Capability for multiplex analysis and effective retrieval of total protein and nucleic acid for mechanism of action and biomarker studies post drug treatment;
- Demonstration that Alvetex Scaffold represents an easy to use, effective and reproducible alternative to the soft agar assay.
General Methods
1 . Preparation of Alvetex Scaffold 96-well plate format
- The 96-well plate was removed from the blister pack within a sterile environment.
- Approximately 100 μl of 70% ethanol was added to each well to prepare the scaffold.
- The ethanol solution was carefully aspirated and 200 μl of appropriate culture medium or PBS was immediately added to wash the scaffold. (This step was performed immediately after the addition of the 70% ethanol because evaporation of the ethanol solution may lead to the scaffold drying out).
- The final wash medium was carefully aspirated. 90 μl of cell culture medium (same as used in the wash step) was added into each well of the plates with liquid handler and placed in incubator 37°C with 7.5 % CO2 until ready for use.
N.B. Care was taken at all times not to touch the Alvetex Scaffold with the pipette tip.
N.B. Hydration with 70 % ethanol, subsequent washing, and addition culture medium were all steps that could be readily performed using an automated liquid handling workstation.
2. Preparation of immortalized cell lines and primary tumor cell (patient-derived xenograft tumor cells) suspensions
- Immortalized cell lines: Cell lines established from the patient-derived xenografts were maintained in culture medium, harvested, counted and plated on the Alvetex Scaffold 96-well plates during the assay procedure. The cell lines were maintained in the culture for a maximum of 12 passages before a new stock was thawed.
- Primary tumor cells (patient-derived xenograft cell suspensions): Cell suspensions from the xenograft material were prepared by enzymatic digestion, rapidly frozen and stored in liquid nitrogen.
- Cells were recovered from cryopreservation for direct seeding into the 96-well plates for drug activity measurement. The seeding densities for primary cells were adjusted based on growth characteristics of each particular tumor type and initial feasibility studies. 50 μl of cell suspension consisting of 7.5 × 104 to 1.5 × 105 cells per ml was seeded per well in 96-well plates with a dispenser. The incubation for each tumor cell suspension was based on the colony formation status of the cells. During this time it was not necessary to change the culture medium for these short-term studies.
3. Addition of drugs and dispensing of compounds
- All cell cultures were maintained overnight at 7.5 % CO2 and 37°C prior to drug addition. Preparation of the compounds was performed on a Tecan EVO 200 automated platform. Compounds were administered directly to the cultures using the MCA component of TECAN EVO 200. The plates were incubated for up to 7 days before being processed for the end point measurement assay.
4. End point measurement assay (ATP-based CellTiter-Glo® assay)
- At the end of the incubation period, the CellTiter-Glo® assay (Promega) was performed. The reagent was dispensed with fast speed into each well of the culture plate with the multi dispenser MCA 96 component of Tecan EVO 200 robot. 130 μl of reagent was dispensed to every 150 μl of cell culture medium per well.
- The plates were subsequently transferred to the Tecan EVO 200 platform and the luminescence signal measured using a Perkin Elmer Envision multimode plate reader. The increased dispensing speed enabled a uniform diffusion of the CellTiter-Glo® assay into the wells and therefore no additional reagent mixing steps were required.
5. Western blot analysis
- Cells were plated and cultured over night as described. After 24 hours the cells were treated (cMET-inhibitor and EGFRinhibitor) and incubated for defined periods (as specified in the methods). At the end of incubation, the culture medium was removed and cells were washed once with PBS. Lysis buffer (Tris pH 7.4, NaCl, EDTA, EGTA, Na4P2O7, NP-40, deoxycholate, glycerol, protease and phosphatase inhibitors) was added to the wells and incubated at 4°C for 30-45 min. After incubation the cell lysates were removed from the wells and transferred to a new plate to spin down the debris. Total protein concentration was determined with BioRad DC Protein Assay. Samples were denatured at 95°C with LDS sample buffer. The total yield of whole cell protein extract per well was approximately 20 to 60 μg. 15 μg (total volume of 30 μl) of whole-cell protein extract was resolved and transferred to PVDF membranes. The resulting blots were saturated/ blocked/ equilibrated in milk, incubated overnight at 4°C with specific primary cMET antibody (Cell Signalling Technology). Detection of GAPDH was used as a loading control. After washing, the membranes were incubated with the appropriate horseradish peroxidase-conjugated secondary antibody (goat anti-rabbit IgG H+L-HRP, Biorad). After further washing, bands were detected using a Pierce Western Blotting Substrate kit (Thermo Scientific) and images captured on a Image Quant LAS 4000 (GE Healthcare).
Results
An introduction to Alvetex Scaffold and the 96-well plate format
Alvetex Scaffold is manufactured from inert polystyrene and is presented as a 200μm thick membrane. Figure 1A shows a sample of the material in transverse section and reveals detail of its highly porous structure. Alvetex Scaffold is 90 % porous and consists of voids (36-40 μm) and interconnecting windows (12-14 μm) creating a 3D space into which cells can grow, proliferate and function. For the 96-well plate format (Figure 1B), Alvetex Scaffold is produced as a sheet that is subsequently welded between the wells and baseplate. Alvetex Scaffold is firmly attached to the baseplate at the bottom of each well and therefore receives cells, and is exposed to the culture medium, from above. Alvetex Scaffold 96-well plates have the same industrial standard dimensions as conventional 2D 96-well plates and are fully compatible with automated work stations and liquid handling robots (Figure 1C).
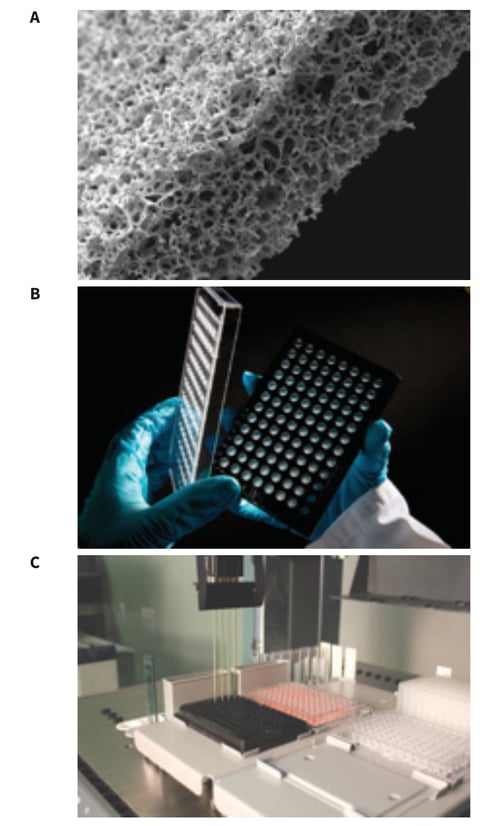 Figure 1: Alvetex Scaffold 96-well plate: (A) Scanning electron micrograph showing the structure of an Alvetex Scaffold membrane in cross-section. Note the highly porous architecture of the polystyrene based material; (B) Alvetex Scaffold 96-well plate product; (C) Automated robotic workstation housing an Alvetex Scaffold 96-well plate with a multichannel pipette head enabling high throughput liquid handling capability.
Figure 1: Alvetex Scaffold 96-well plate: (A) Scanning electron micrograph showing the structure of an Alvetex Scaffold membrane in cross-section. Note the highly porous architecture of the polystyrene based material; (B) Alvetex Scaffold 96-well plate product; (C) Automated robotic workstation housing an Alvetex Scaffold 96-well plate with a multichannel pipette head enabling high throughput liquid handling capability.
Enabling 3D cancer cell culture in the Alvetex Scaffold 96-well plate
A substantial body of evidence exists showing 3D growth of cells in Alvetex Scaffold products for a broad range of alternative applications (for further details consult our website at Alvetex Protocols). Similarly, and for the purposes of demonstration only, we provide direct evidence of cells growing in Alvetex Scaffold when presented in the 96-well plate format. Figure 2 shows data for two different cell types growing for up to a week in an Alvetex Scaffold 96-well plate. It should be noted that not all cell types behave in the same way. Because cells differ in their size, proliferation rate and migratory ability, different cell types will grow and populate the scaffold in different ways. Some cell types will completely penetrate the scaffold membrane whilst others will reside primarily in the upper levels of the material. In both cases the cells acquire and maintain a natural 3D architecture and phenotype. The data presented in Figure 2 show growth patterns for two different well-known cancer cell lines. LN-229 glioblastoma cells rapidly enter the scaffold and show a more diffuse cell distribution throughout the scaffold (Figure 2A). In comparison, SW620 colorectal adeno-carcinoma cells are more restricted to the upper third of the scaffold and grow more densely as clumps of cells (Figure 2B,C). The individual 3D shape of the cells is also noticeably different between the two cell types: LN-229 cells resemble a more fibroblastic phenotype, whereas SW620 cells are smaller and rounded. (It should be noted that we do not recommend routine histological analysis of 3D cultures using the Alvetex Scaffold 96-well plate format. If detailed information concerning the structural phenotype of the cell culture is required, we recommend using the Alvetex Scaffold well insert format. Full details about cell visualisation and accompanying protocols are available online at Alvetex Protocols).
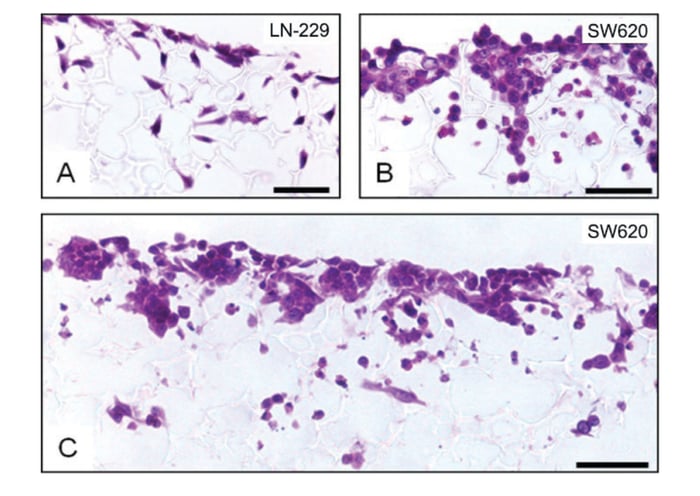 Figure 2: Histological analysis of LN-229 glioblastoma (A) and SW620 colorectal adenocarcinoma (B, C) cells cultured for 3 (A) or 7 (B, C) days in 3D using an Alvetex Scaffold 96-well plate. The lower magnification image (C) shows that cells primarily populate the upper 50 % of the scaffold and are distributed relatively evenly across the membrane. Scale bars: (A, B) 25 μm; (C) 50 μm.
Figure 2: Histological analysis of LN-229 glioblastoma (A) and SW620 colorectal adenocarcinoma (B, C) cells cultured for 3 (A) or 7 (B, C) days in 3D using an Alvetex Scaffold 96-well plate. The lower magnification image (C) shows that cells primarily populate the upper 50 % of the scaffold and are distributed relatively evenly across the membrane. Scale bars: (A, B) 25 μm; (C) 50 μm.
Luminescence-based cell viability assay using the Alvetex Scaffold 96-well plate
The growth and expansion of cancer cell populations in 3D culture can be monitored in Alvetex Scaffold 96-well plates using standard commercial assay kits. In this study we used Promega’s CellTiter-Glo® luminescent cell viability assay. CellTiter-Glo® is a homogeneous method of determining the number of viable cells in culture based on quantitation of the ATP present, an indicator of metabolically active cells. The CellTiter-Glo® Assay is designed for use with multi-well formats, making it ideal for automated high-throughput screening, cell proliferation and cytotoxicity assays. The assay generates a “glow-type” luminescent signal with an extended half-life of up to 5 hours which eliminates the need to use reagent injectors and provides flexibility for continuous or batch mode processing of multiple plates. The homogeneous format avoids errors that may be introduced by other ATP measurement methods that require multiple steps.
A routine dose response experiment was performed using single cell suspensions prepared from patient-derived non-small cell lung cancer (NSCLC) xenografts cultured in Alvetex Scaffold 96-well plates in the presence of the topoisomerase I inhibitor, SN38. This potential anti-cancer agent is understood to arrest cells in S- and G2-phases of the cell cycle, and induce apoptosis of cancer cells via activation of caspase 3 and PARP (Maurya et al. 2011; Voigt et al. 1998). Specifically it has been found to induce these effects in multiple lung carcinoma cell lines indicating its potential value as an effective therapeutic agent against lung cancer (Maurya et al. 2011). To assess cytotoxicity, a dose response curve (10 concentrations, half log dilution steps) for SN38 was generated using the NSCLC cells as described above (Figure 3). Cells were exposed to the compound for 7 days (post drug application) followed by assessment of drug response using the CellTiter-Glo® assay. The results show a typical dose response profile and demonstrate the effectiveness of SN38 in reducing the viability of these cells.
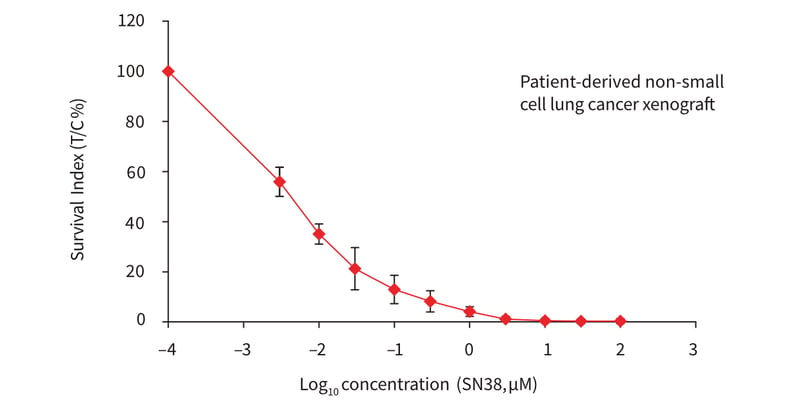 Figure 3: Plot showing a dose response curve for cell suspensions from a patient-derived non-small cell lung cancer xenograft, treated with SN38 (topoisomerase I inhibitor) in Alvetex Scaffold 96-well plates. A total of 10 point concentrations with an increment of half log dilution steps were tested. The cells were incubated further for 7 days in the presence of the inhibitor and cell viability was subsequently measured with the ATP-based CellTiter-Glo® luminescence assay. The x-axis represents concen-tration in BM and the Survival Indices (treated (T) versus control (C), T/C %) are shown on the y-axis. Data represent mean ± SD, n = 4.
Figure 3: Plot showing a dose response curve for cell suspensions from a patient-derived non-small cell lung cancer xenograft, treated with SN38 (topoisomerase I inhibitor) in Alvetex Scaffold 96-well plates. A total of 10 point concentrations with an increment of half log dilution steps were tested. The cells were incubated further for 7 days in the presence of the inhibitor and cell viability was subsequently measured with the ATP-based CellTiter-Glo® luminescence assay. The x-axis represents concen-tration in BM and the Survival Indices (treated (T) versus control (C), T/C %) are shown on the y-axis. Data represent mean ± SD, n = 4.
In general, these data demonstrate the compatibility of Alvetex Scaffold 3D cell culture technology for this type of medium to high throughput cytotoxicity assay, thereby representing an attractive alternative to the routine soft agar based pre-clinical drug profiling assays. In this context it is important to recognise that soft agar growth assays have been used for decades. Consequently, huge historical data sets have been built up that contain valuable information about compound activities on different cancer cell types. In order to demonstrate that the basic data generated from this new method is consistent with that previously observed, we compared the cytotoxicity of SN38 and erlotinib. Erlotinib hydrochloride (trade name Tarceva®, marketed by Roche) is a known reversible tyrosine kinase inhibitor (anti-EGFR, Rocha-Lima CM & Raez LE). EGFR is often highly expressed and occasionally mutated in various forms of cancer (Bulgaru et al. 2003). Erlotinib has been widely used to treat patients with non-small cell lung cancer as a first line or second line therapy (Román Pérez-Soler, 2004; Fiala O, et al. 2013), pancreatic cancer (Kelly and Ko, 2008) and several other types of cancers including CRC. med plates containing soft agar (Figure 4). After 24 hours initial growth, cells were exposed for 7 days to ten concentrations of SN38 or erlotinib prior to analysis of cell viability using the CellTiter-Glo® assay. All conditions for cell handling, drug addition and analysis were identical. The profiles of the dose response curves were observed to be similar. Overall, the data acquired from the Alvetex Scaffold format were reproducible and correlated well with data produced in the soft agar assay format.
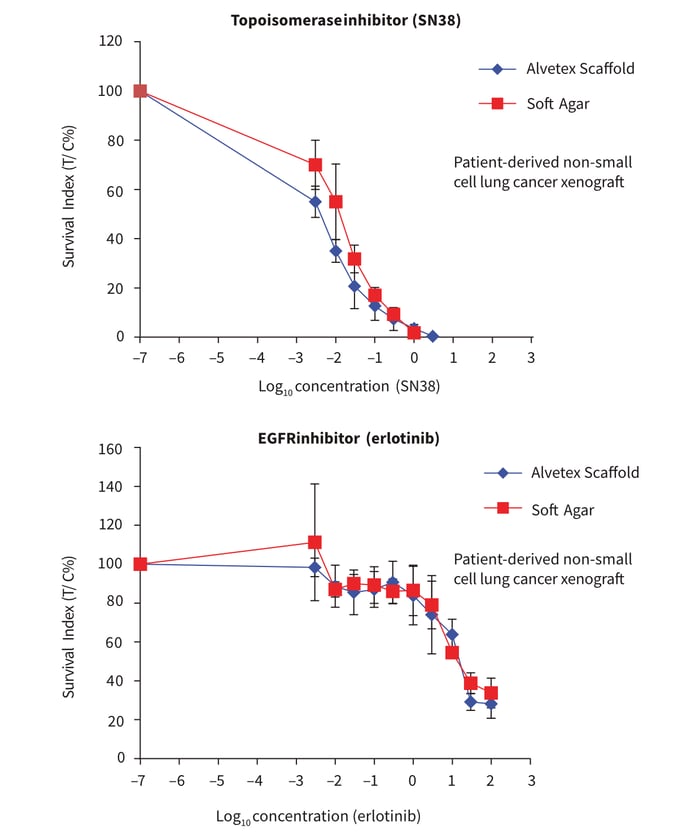 Figure 4. Comparison of 3D culture methods using Alvetex Scaffold well plates and a conventional soft agar assay. Cancer cells derived from patient-derived non-small cell lung cancer xenografts were seeded in Alvetex Scaffold 96-well plates and 96-well flat bottom plates containing soft agar. All conditions were kept identical for both types of plate format. The plates were initially incubated for 24 hours prior to treatment with com-pounds: Topoisomerase I inhibitor (SN38) (A) or EGFR inhibitor (erlotinib) (B). Ten point concentrations were used with an increment of half log dilution steps between each concentration. After 7 days drug treatment, cell viability was assessed using the ATP-based CellTiter-Glo® luminescence assay. The x-axis represents concentration in μM and the Survival Indices (T/C %) are shown on the y-axis. Data represented as mean ± SD, n = 2-4.
Figure 4. Comparison of 3D culture methods using Alvetex Scaffold well plates and a conventional soft agar assay. Cancer cells derived from patient-derived non-small cell lung cancer xenografts were seeded in Alvetex Scaffold 96-well plates and 96-well flat bottom plates containing soft agar. All conditions were kept identical for both types of plate format. The plates were initially incubated for 24 hours prior to treatment with com-pounds: Topoisomerase I inhibitor (SN38) (A) or EGFR inhibitor (erlotinib) (B). Ten point concentrations were used with an increment of half log dilution steps between each concentration. After 7 days drug treatment, cell viability was assessed using the ATP-based CellTiter-Glo® luminescence assay. The x-axis represents concentration in μM and the Survival Indices (T/C %) are shown on the y-axis. Data represented as mean ± SD, n = 2-4.
Performing drug combination cancer cell cytotoxicity assays using the Alvetex Scaffold 96-well plate
New combination strategies have already been proven to significantly increase the efficacy of treatment regimens in cancer therapy. Cell based assays are a useful tool to evaluate rational and exploratory combinations in vitro in a pre-clinical phase. However, investigation of combina-tion therapies is cumbersome on account of the number of data points to be acquired. To enable testing of a large number of rational combinations under in vitro and ex vivo conditions we developed a “combination screening approach” using a fixed ratio layout using only 5 concentrations of the combination partners. This new methodology is feasible for medium to high throughput combination assays and was adapted to the Alvetex Scaffold format.
Combination therapy was investigated using an Oncotest proprietary colon cancer cell line established from a patient-derived colorectal cancer (CRC) xenograft. Here, two drugs approved for treatment of CRC, ispinesib and erlotinib, were applied in monotherapy and in com-bination. Ispinesib is a small molecule inhibitor of Kinesin spindle protein (Kspi) that acts to suppress mitosis resulting in growth inhibition in several types of cancer (Purcell et al. 2011). In recent years, the potential of erlotinib for advanced metastatic CRCs has been investigated mainly in new combination strategies (Weickhardt et al. 2012). Ispinesib was selected for a combination with erlotinib based on Oncotest internal reference data (data not shown).
For comparison of 2D and 3D cell culture approaches, CRC cells were grown in either conventional 2D or Alvetex Scaffold 3D 96-well culture plates. After 24 hours initial growth, cells were exposed to five concentrations (log dilution steps) of the test compounds over a four day incubation period prior to analysis of cell viability using the CellTiter-Glo® assay. Treatment with erlotinib in monotherapy did not result in any substantial loss in cell viability in cultures grown in conventional 2D plates (Figure 5A). However, cells cultured on Alvetex Scaffold 3D 96-well culture plates were observed to be relatively more sensitive to erlotinib at the highest concentration (Figure 5B) as compared to the cells growing in mono-layer. This discrimination was possible even in the reduced combination screening format. Nevertheless, there is evidence towards an enhanced drug activity in combination in both cell culture methods. These data further demonstrate the suitability of Alvetex Scaffold 3D 96-well culture plates for cytotoxicity assays and drug combination studies.
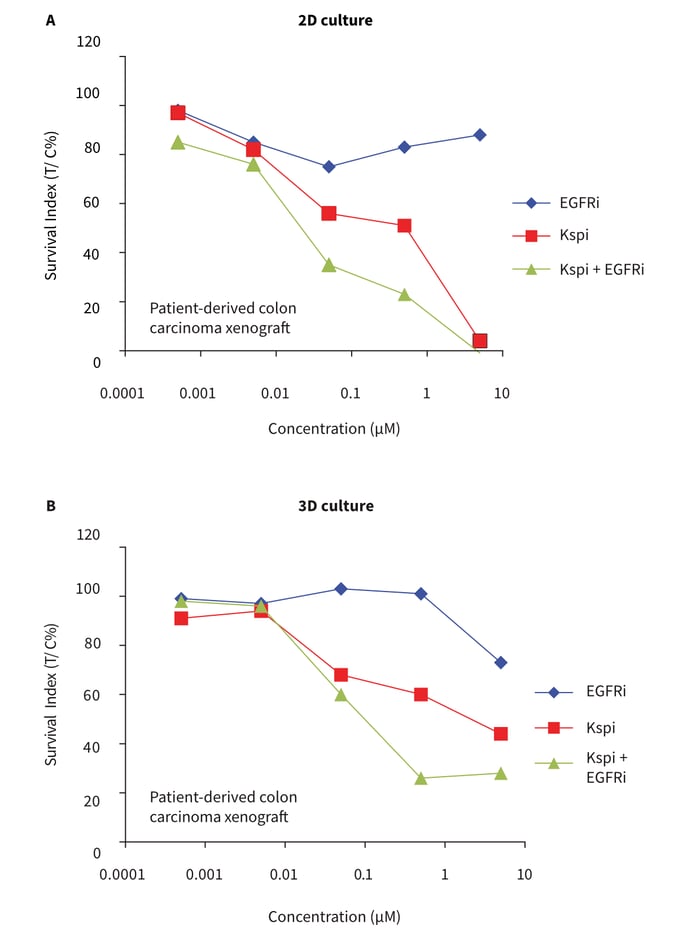 Figure 5: Five-point concentration drug combination assay and comparison of 2D vs 3D cell culture formats. Cells from a colon carcinoma cell line were seeded in either conventional 96-well flat bottom plates (A, standard 2D cell culture format) or in Alvetex Scaffold 96-well plates (B, 3D cell culture format). Cells were exposed to the targeted drugs, ispinesib (Kspi) and EGFR inhibitor erlotinib (EGFRi) alone and in combination over ×5 log-dilution step concentrations. Cultures were maintained for 4 days during which no media changes were per-formed. Subsequently, cell viability was measured with the ATP-based CellTiter-Glo® luminescence assay. The x-axis represents concentration in BM and the Survival Indices (T/C %) are shown on the y-axis.
Figure 5: Five-point concentration drug combination assay and comparison of 2D vs 3D cell culture formats. Cells from a colon carcinoma cell line were seeded in either conventional 96-well flat bottom plates (A, standard 2D cell culture format) or in Alvetex Scaffold 96-well plates (B, 3D cell culture format). Cells were exposed to the targeted drugs, ispinesib (Kspi) and EGFR inhibitor erlotinib (EGFRi) alone and in combination over ×5 log-dilution step concentrations. Cultures were maintained for 4 days during which no media changes were per-formed. Subsequently, cell viability was measured with the ATP-based CellTiter-Glo® luminescence assay. The x-axis represents concentration in BM and the Survival Indices (T/C %) are shown on the y-axis.
Multiplex analysis – using the Alvetex Scaffold 96-well plate for combined 3D culture, cytotoxicity and expression analyses
Over the last years several targeted agents have been approved as chemotherapeutics; therefore, it is of great interest to examine the changes of expression patterns of specific genes and target proteins when studying the cytotoxic effects of these compounds. Such information is of great value to understand in detail the molecular mechanisms by which the test compounds act. We therefore tested the ability to isolate (i) total protein (Figure 5) and (ii) nucleic acid from 3D cancer cell cultures grown using Alvetex Scaffold 96-well plates after drug treatment.
Retrieval of biological material was tested in the course of a combination experiment. Single cell suspensions prepared from NSCLC xenografts were seeded into Alvetex Scaffold 3D 96-well culture plates. After 24 hours initial growth, cells were exposed for 7 days to five concentrations of a cMET kinase inhibitor (cMETi) and erlotinib, either individually or in combination. The receptor tyrosine kinase cMET stimulates cell invasion and protection from apoptosis. Dysregulated activity of cMET is associated with a wide variety of different cancers, such as lung carcinomas. The dose response data indicate that cell viability was maintained at high levels in the presence of either compound when used in monotherapy up to concentrations of 1-10 μM. At higher concentrations, moderate levels of cytotoxicity were observed (Figure 6A). When the drugs were combined, however, the toxic effects on cell viability were significantly greater and detectable at lower compound concentrations, indicating enhanced activity of the compounds when tested in combination.
Total protein was isolated from each well of the Alvetex Scaffold 96-well plate in preparation for protein expression analyses. Approximately 20-60 mg of total protein at a maximum concentration of about 1 mg/ml was isolated from one well. In a subsequent Western blot analysis (Figure 6B), the phosphorylation status (P activity status) of cMET was assessed to monitor the efficacy of the cMETi at molecular level. In order to exclude, (as far as possible) a misinterpretation of the results by changes in protein expression related to reduced cell viability or even cell death induced by the compounds, only samples representing the first three concentrations (C1 -C3) were processed for Western blot analysis. The data clearly demonstrate that NSCLC cells growing in 3D culture are susceptible to cMet inhibition, as indicated by a decreased phospho-cMET signal at increasing compound concentrations. Changes in phosphorylated cMET were thus already apparent and detectable before any significant influence to cell viability was observed. These data underline the value of multiplexing where a single sample can be analysed by multiple methods to provide additional useful information. Importantly, Alvetex Scaffold 3D 96-well culture plates are compatible with many such methods. In this context it is noteworthy that sufficient amounts of mRNA could also be recovered for identification of biomarkers post drug treatment when using techniques such as qPCR (data not shown).
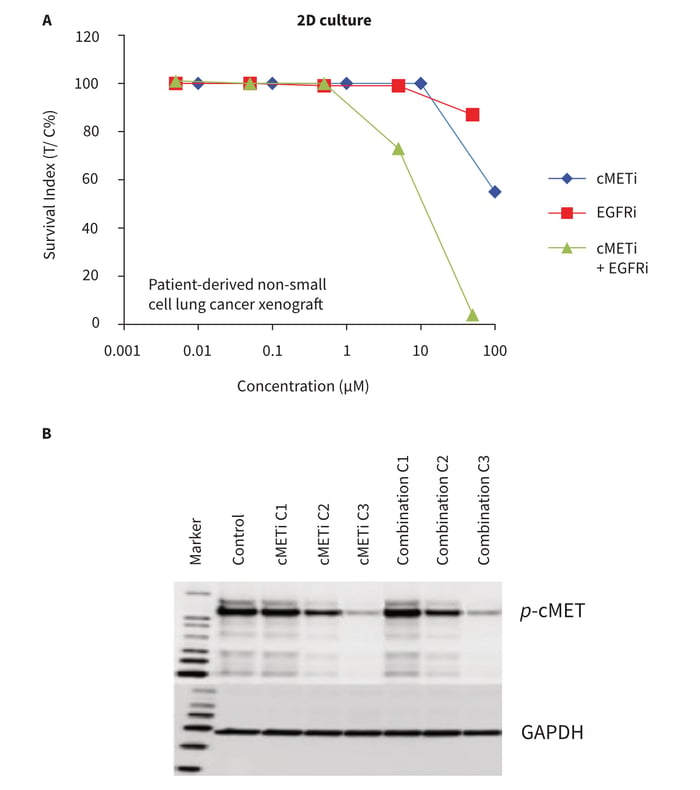 Figure 6: Drug combination study in conjunction with subsequent biomarker expression analysis post treatment. NSCLC cells derived from a PDX were grown in Alvetex Scaffold 96-well plates and treated in mono-therapy or in combination with cMet kinase inhibitor (cMETi) and/or EGFR inhibitor (EGFRi). After 7 days, cell viability was measured with the ATP-based CellTiter-Glo® luminescence assay (A). The data show the outcome of a typical experiment in duplicate. The x-axis represents concentration in BM and the Survival Indices (T/C%) are shown on the y-axis. Protein lysates were recovered after drug treatment and analysed for phosphorylated cMet levels by Western blot analysis (B). Samples: cMet inhibitor alone (cMETi); combination (cMeti and EGFRi); at the three lowest concentrations (C1-C3, see A); GAPDH was used as a loading control. Note that no effect on cell viability was observed at drug concentrations C2 and C3 but changes in the phosphorylation status of cMET were already apparent and detectable.
Figure 6: Drug combination study in conjunction with subsequent biomarker expression analysis post treatment. NSCLC cells derived from a PDX were grown in Alvetex Scaffold 96-well plates and treated in mono-therapy or in combination with cMet kinase inhibitor (cMETi) and/or EGFR inhibitor (EGFRi). After 7 days, cell viability was measured with the ATP-based CellTiter-Glo® luminescence assay (A). The data show the outcome of a typical experiment in duplicate. The x-axis represents concentration in BM and the Survival Indices (T/C%) are shown on the y-axis. Protein lysates were recovered after drug treatment and analysed for phosphorylated cMet levels by Western blot analysis (B). Samples: cMet inhibitor alone (cMETi); combination (cMeti and EGFRi); at the three lowest concentrations (C1-C3, see A); GAPDH was used as a loading control. Note that no effect on cell viability was observed at drug concentrations C2 and C3 but changes in the phosphorylation status of cMET were already apparent and detectable.
The recovery of protein and/or nucleic acids from the Alvetex Scaffold 3D 96-well culture plates enables greater versatility in understanding the underlying molecular mechanisms in a more physiologically relevant 3D culture model compared to conventional 2D 96-well culture plates.
Practical benefits of using Alvetex Scaffold 3D culture technology over the existing soft agar growth assay
The formation of colonies in soft agar has been the primary method of monitoring anchorage-independent growth for decades and is well established and accepted. However, this approach is laborious and requires significant preparation time. With respect to targeted agents, it is essential to perform multiplex analysis on in vitro or ex vivo-derived samples. The soft agar approach is challenging when subsequent analysis of protein and nucleic acid expression is required. The process to extract and isolate such samples is complicated by the presence of the semisolid support that may interfere with the procedures used, thus influencing the outcome of the analysis.
We have demonstrated herein the application of Alvetex Scaffold technology for 3D cell culture and cytotoxicity assays used for pre-clinical profiling. Additionally, this new assay format eases subsequent multiplex analysis within a single approach. This is mainly based on the direct accessibility of the cells growing in Alvetex Scaffold which enables a simple and clean isolation of the cells or biological material of interest. In the context of molecular analyses it is noteworthy that Alvetex Scaffold is made from inert polystyrene, and given the physical properties of the material, it does not interfere with downstream assays. Alvetex Scaffold is hydrophobic and cells may not attach strongly to the scaffold but rather form 3D contacts with adjacent cells, creating cell aggregates and rudimentary 3D tissue-like structures. Cells that naturally form colonies do so within the voids and on the surface of Alvetex Scaffold. Drugs therefore have direct access to the cells in the highly porous scaffold and their diffusion is not limited by a semisolid support. In a practical sense, Alvetex Scaffold is suitable for automated platforms and easier to use than the soft agar method. In certain applications therefore, Alvetex Scaffold represents a credible alternative to the soft agar approach, enabling straightforward and routine 3D cell culture, assessment of cytotoxicity and subsequent multiplex analysis.
References
- Maurya DK, Ayuzawa R, Doi C, et al. Topoisomerase I inhibitor SN-38 effectively attenuates growth of human non-small cell lung cancer cell lines in vitro and in vivo. J Environ Pathol Toxicol Oncol (2011) 30:1-10.
- Voigt W, Matsui S, Yin MB, et al. Topoisomerase-I inhibitor SN-38 can induce DNA damage and chromosomal aberrations independent from DNA synthesis. Anticancer Res (1998) 18:3499-3505.
- Rocha-Lima CM, Raez LE. Erlotinib (Tarceva) for the Treatment of Non–Small-Cell Lung Cancer and Pancreatic Cancer. Pharmacy and Therapeutics (2009) 34: 554-564.
- Bulgaru AM, Mani S, Goel et al. Erlotinib (Tarceva®): a promising drug targeting epidermal growth factor receptor tyrosine kinase. Expert Rev Antican Ther (2003) 3:269-279.
- Román Pérez-Soler, MD, et al. “Selected Highlights”. Lung Cancer Front (2004) 22:3238–3247.
- Fiala O, Pesek M, Finek J, et al. Second line treatment in advanced non-small cell lung cancer (NSCLC): Comparison of efficacy of Erlotinib and chemo-therapy. Neoplasma (2013) 60:129-134.
- Kelley K, Ko A. Erlotinib in the treatment of advanced pancreatic cancer. Biologics (2008) 2:83–95.
- Purcell J, Davis J, Reddy M, et al. Activity of the Kinesin Spindle Protein Inhibitor Ispinesib (SB-71 5992) in Models of Breast Cancer. Clin Cancer Res (2010) 16:566–576.
- Weickhardt AJ, Chong G, Gebski V, et al. Dual Targeting of the Epidermal Growth Factor Receptor Using the Combination of Cetuximab and Erlotinib: Preclinical Evaluation and Results of the Phase II DUX Study in Chemotherapy-Refractory, Advanced Colorectal Cancer. J Clin Oncol (2012) 30:1505-1512.