Total Protein Extraction from Cells Cultured in Alvetex® Scaffold in 3D
● Download this protocol as a PDF (1.1 MB)
1. Introduction
The following protocol outlines a method for the extraction of total protein from cells cultured in Alvetex Scaffold. Example data was obtained using this protocol to extract protein from HepG2 hepatocytes cultured in Alvetex Scaffold for 7 days in 6-well inserts (AVP004) in Well Insert Holder in Deep Petri Dish (AVP015) format.
2. Method
- Prepare lysis buffer: 1 % v/v Igepal® CA-630 (Sigma, Product No. I3021), 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM MgCl2, and Complete Mini Protease Inhibitor Cocktail (Roche Diagnostics, 11 836 153 001).
Note: The Protease Inhibitor Cocktail is available in two forms, with or without EDTA lysate, see manufacturer instructions for recommendation.
- Remove Alvetex Scaffold disc from culture format (e.g. well insert or 12-well multiwall plate) and wash by gentle immersion in PBS (e.g. in a Petri dish). Transfer into a 1.5 mL tube.
- Add 1 mL of lysis buffer and vortex for 10 seconds. Place on ice for 30 minutes with vortexing (10 sec.) every 5 minutes.
- Clarify lysates by centrifugation at approx 15,000 rpm for 3 minutes and place supernatant into a fresh tube.
- The samples can be frozen at this stage (e.g. at –80 °C), for use at a later date.
- Determine total protein content of samples using a protein determination assay (e.g. Bio-Rad
protein assay).
3. Example: Extraction of Total Protein from HepG2 Hepatocytes Cultured in Alvetex Scaffold in 3D Cell Culture
HepG2 cells (ATCC, HB-8065) were routinely maintained in T-75 flasks. HepG2 complete media consisted of: MEM media (Gibco, 21090) supplemented with 10 % v/v FBS, 2 mM L-glutamine and 100 U/mL Penicillin/ Streptomycin. Cells were seeded onto Alvetex Scaffold discs in 6-well inserts (AVP004-3) in well insert holders in deep Petri dishes (Product No. AVP015), at a density of 1 × 106 cells in 150 μL media per insert. After settling for 3 hours in an incubator (5 % CO2, 37°C) complete media was carefully added (70 mL per Petri dish). Cultures were maintained for 7 days, with a single media change on day 5. After 7 days, well inserts were dismantled and cultures were processed according to the protocol described above.
3.1. Protein analysis results
Determination of relative protein concentration was made using Protein Assay Reagent (Bio-Rad, 500-0006). A dilution series of BSA was used for the generation of a standard curve (Figure 1. inset). Approximately 1 milligram of protein was isolated per disc (Table 1).
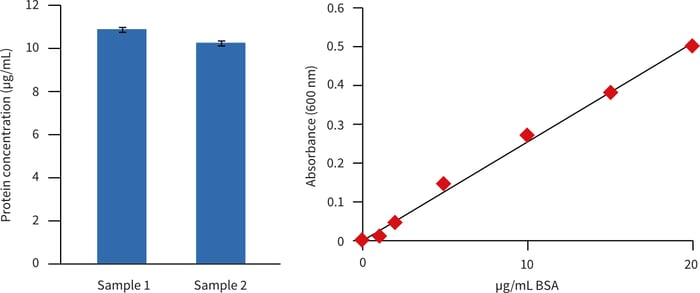
Figure 1. Estimation of protein concentration (relative to BSA) in extracts from Alvetex Scaffold cultures of HepG2 hepatocytes. Extracts were assayed at a dilution of 1 in 100. Data from two biological replicate cultures are shown; each sample was read in triplicate.
| Sample 1 | Sample 2 | |
|---|---|---|
| Protein yield (mg/mL) | 1.08 (± 0.01) | 1.02 (± 0.01) |
Table 1. Approximate protein yields in extracts from Alvetex Scaffold cultures of HepG2 hepatocytes, using BSA as a standard reference. Values shown represent the mean of triplicate readings per sample (± SD).
Protein extracts were also analysed by SDS-polyacrylamide gel electrophoresis and western blotting (Figure 2). Protein extracts from two biological replicate cultures produced well-resolved and highly reproducible SDS-PAGE profiles as determined via Coomassie Brilliant Blue R staining. Western blotting demonstrated that the constitutive protein β-actin was both intact and equally loaded in both sample lanes.
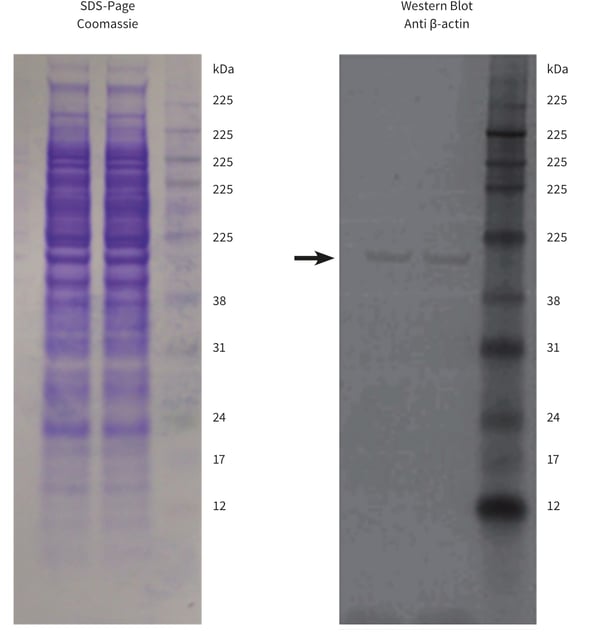
Figure 2. SDS-polyacrylamide gel electrophoresis and western blot analysis of protein extracts from Alvetex Scaffold cultures of HepG2 hepatocytes. Polyacrylamide gels (NuPAGE 10% Bis-Tris, Invitrogen, NP0302) were loaded with 20 µg protein. Resolved gels were stained with Commassie Brilliant Blue R (left) or blotted to PVDF membrane for western analysis (right). The western blot was probed for the constitutive protein β-actin (arrow). Data from two biological replicate cultures are shown.