XTT Cell Viability Assay for Alvetex® Scaffold 96 Well Plate Format
● Download this protocol as a PDF (0.1 MB)
1. Introduction
This chromogenic assay involves the reduction of XTT (2,3-Bis-(2-Methoxy-4-Nitro-5-Sulfophenyl)- 2H-Tetrazolium-5-Carboxanilide) to form an orange coloured formazan derivative. Although mitochondrial oxidoreductases are thought to contribute significantly to this conversion, actual reduction of XTT is believed to occur at the extracellular surface of the plasma membrane, where the sensitivity of the assay can be enhanced by addition of the intermediate electron carrier, PMS (N-methyl dibenzopyrazine methyl sulphate). This results in the colour change taking place directly in the culture medium without cellular import of the XTT substrate. Therefore, the XTT reagent is suitable for use in real time metabolic viability assays. It is recommended that for each cell type the linear relationship between cell number and signal produced should be first established in order to investigate the limits of the assay.
2. Method
- This is an example protocol, which may require further optimisation depending on the rate of cell growth of a particular cell type on Alvetex Scaffold.
- Prepare activated XTT solution by adding 0.1 mL Activation Reagent per 5.0 mL XTT reagent according to the manufacturer’s instructions (ATCC, 30-1011K).
- Withdraw media from each well of the Alvetex Scaffold 96 well plate and replace with fresh media mixed 2:1 with Activated XTT reagent (100 µL media plus 50 µL Activated XTT reagent = 150 µL total volume per well). Alternatively, leave 100 µL existing media in place and add 50 µL of Activated XTT reagent directly to each well.
- Include a blank control well (no cells) containing 100 μL medium plus 50 μL activated XTT solution only.
- Incubate plate for 1-2 hours at 37 °C with CO2 levels appropriate to cell type.
- Agitate sample by pipetting up and down and transfer 100 μL from each well directly into a clean standard 96 well plate.
- Using a microtiter plate reader measure the absorbance of all wells (a.) between 450-500 nm followed by (b.) measurement between 630-690 nm for non-specific absorbance. In the example below, readings were taken 450 nm and 630 nm.
- Calculate specific absorbance as follows:
Specific Absorbance = A450nm (sample) – A450nm (blank) – A630nm (sample)
3. Example data
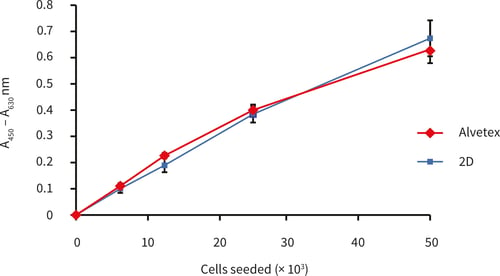
Figure 1. XTT assay of HepG2 cells cultured for 24 hours in Alvetex Scaffold in 96 well plate format (n = 3, mean ± SD). Good linearity was observed between cell number and signal produced, up to a seeding density of 50,000 cells per well. Data obtained was comparable between Alvetex Scaffold and standard 2D plates.