What is digital droplet PCR and how can it improve stem cell therapies?

To accurately quantify the number of undifferentiated cells in a cell therapy product, scientists have developed a ddPCR-based assay called Sistem. The following article explains how this assay can be used to make cell and gene therapies safer.
What is digital droplet PCR?
Droplet digital PCR (ddPCR) is a screening method that is much more precise than conventional PCR (qPCR). With qPCR, you only get a single fluorescent measurement - perhaps five or six if you're trying to multiplex it. But with ddPCR, we can achieve up to 20,000 fluorescent measurements in a single reaction.
This increased precision, robustness, and sensitivity means we can detecting lower copy numbers compared with qPCR. So if you're trying to find individual cells in a population, it's very useful. Compared to other PCR types it's also relatively insensitive to PCR inhibition and can achieve absolute quantification. So there is no need to worry about normalizing the target, having the ideal calibrated gene for the sample, or running a standard in order to carry out absolute quantitation.
How does digital droplet PCR work?
In ddPCR, the sample is partitioned into 20,000 droplets of a defined volume, which distributes the RNA or DNA randomly amongst the droplets. Following amplification, the droplet reader is sensitive enough to detect the fluorescence signal in every individual droplet. It measures whether the droplet contains the nucleic acid of interest and then performs absolute quantification of all the molecules in the reaction. Performing this absolute quantification is a key advantage of ddPCR, which uses Poisson distribution to correct for multiple target molecules per droplet.
How can ddPCR be used to make stem cell therapies safer?
Induced pluripotent stem cell (iPSC) therapies show unrivaled potential in treating incurable conditions, such as Spinocrebrall Ataxia and Parkinson's disease. Yet they present a risk of tumorgenicity if undifferentiated cells reside in the final product, so it is essential to quantify the number of undifferentiated cells in a product before human trials.
However, there is currently no standardized way to assess levels of residual iPSCs in cell preparations. Even if the number of residual undifferentiated cells is accurately quantified by a manufacturer, several regulatory bodies request that an independent testing agent validates their results. To complicate matters further, many of the existing tests lack the sensitivity required to detect the low number of undifferentiated cells. And tests that are sensitive enough are usually expensive, time-consuming, and laborious.
This is where ddPCR comes in. By screening for micro RNAs (miRNAs) only found in undifferentiated cells, you can access a sensitive, inexpensive assay that can be used for routine quality control. We developed an assay called Sistem to do just that.
What is SistemPSCCheck?
Sistem uses ddPCR to screen miRNAs that should only be found in undifferentiated cells. A key advantage of this assay is that it identifies the absolute copy number of miRNA targets and how this correlates with the number of undifferentiated cells in the preparation. Using this relationship, Sistem accurately quantifies the number of embryonic stem cells or iPSCs in a preparation, which can contain any differentiated cell type.
Typically, Sistem is used when a client is looking to validate a batch, or if they're trying to validate their own internal method. We receive frozen cell palettes from the client and they receive results within 7-10 business days, depending on the number of cells.
Why use miRNAs?
We were interested in using miRNAs because they maintain stem cell pluripotency, differentiation, and self-renewal. These molecules are also useful from a biomarker diagnostic standpoint because they are stable in most forms; it doesn't matter if they are received as frozen cells or cellular lysate, miRNAs don't degrade compared to other nucleic acids. You can even extract them from tissues preserved in wax, which isn't possible with DNA or RNA.
How did Sistemic develop this ddPCR assay?
SistemPSCCheck was optimized using samples using iPSCs as the undifferentiated cell source, and bone marrow-derived MSCs (BM-MSCs) as the differentiated cell source. This was achieved by either spiking a known number of iPSCs into a background of 1×105 BM-MSCs, or spiking the equivalent concentration of iPSC RNA into a background of BM-MSC RNA.
To determine the biomarkers used to screen samples, we used a miRNA database to identify markers associated with undifferentiated cells. Using multiple microarrays and qPCR reactions, we then identified miRNA targets expressed at high levels in undifferentiated cells and low levels in differentiated cells. We selected the top four biomarkers with high expression in stem cells and low expression in differentiated cell types.
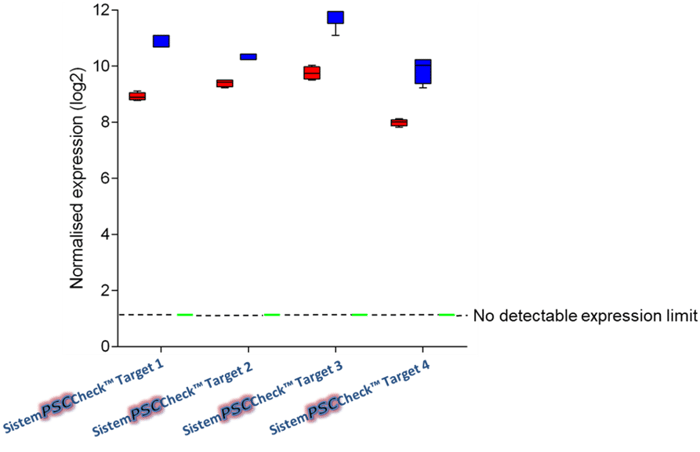 Figure 1: SistemPSCCheck is specific for undifferentiated cells. In this graph here, we have three different groups: iPSCs (red), embryonic stem cells (ESCs, blue), non-pluripotent cells, and terminally differentiated cells (green). While there is a high expression of the markers in the iPSC and ESC groups, the non-pluripotent cells and the differentiated cells do not show any expression.
Figure 1: SistemPSCCheck is specific for undifferentiated cells. In this graph here, we have three different groups: iPSCs (red), embryonic stem cells (ESCs, blue), non-pluripotent cells, and terminally differentiated cells (green). While there is a high expression of the markers in the iPSC and ESC groups, the non-pluripotent cells and the differentiated cells do not show any expression.
How was SistemPSCCheck validated?
Assay performance specification was carried out using a number of tests based on the ICH Q2 guidelines: target specificity, linearity, detection sensitivity, and intra-assay performance, and inter-assay performance.
- Target Assay Specificity
The results of this validation showed that the assays have >99% specificity for their unique miRNA target. There was also no cross-reactivity with other targets, meaning that it's unlikely that one assay will cause another to have a false-positive result.
- Assay Linearity
To test assay linearity, we RNA spiked 1000, 100, 10, or 1 iPSC in a background of 1×105 BM-MSCs. As the concentration of iPSC RNA increased, we saw increased expression across all four biomarkers. This suggests that SistemPPCCheck shows excellent linearity between targets in respect to copy and iPSC number.
- Assay detection sensitivity
Using the cell spike samples, we then tested assay sensitivity for all four targets. This confirmed that we could constantly detect 5-10 iPSCs per 1×105 BM-MSCs compared to the control, however, it was difficult to distinguish 1-2 iPSC. We set the LLQ of the assay between 3-5 iPSCs per 1×105 BM-MSCs.
- Intra-assay performance
Validating intra-assay performance, meant having two different analysts conduct the assay using different equipment, on the same samples, at the same laboratory, on the same day. SistemPPCCheck showed good reproducibility between the analysts, meaning assay performance is not operator dependant.
- Inter-assay performance
Estimating inter-assay analysis required one analyst to conduct the assay on the same samples on consecutive days. Again, we obtained consistent expression levels, suggesting that the assay is reproducible over different days. The final requirement was to have the national measurement laboratory independently validate the assay performance, which showed comparable expression between the analysts for all four targets.
- Diagnostic sensitivity and specificity
After performing the above validation steps, we could finally calculate the diagnostic accuracy of the assay. SistemPPCCheck gave no false-negative results (meaning the assay has 100% sensitivity at 5-10 iPSCs per 1×105 BM-MSCs) and no false-positive results (100% specificity). It is, therefore, a very accurate method for finding low copies of undifferentiated cells in derived cell therapeutics.
A highly sensitive, specific, ddPCR-based assay
SistemPSCCheck has good target specificity for undifferentiated cells. The assays used have 99% specificity for their unique miRNA target and show excellent linearity between target expression and iPSC number/RNA concentration. Regardless of the day, operator, or lab, the assay consistently detects both 5-10 iPSCs per 1×105 BM-MSCs. It is highly reproducible and can be validated in different laboratories if necessary.

Author
Zara Puckrin, BSc
Zara is a GCU graduate who loves minimalism, marketing, and molecular biology. You can contact her on LinkedIn.
Subscribe to receive updates from REPROCELL
Tagged
Stem cell and drug discovery scientists around the world are using REPROCELL’s services and products in their preclinical and clinical research.